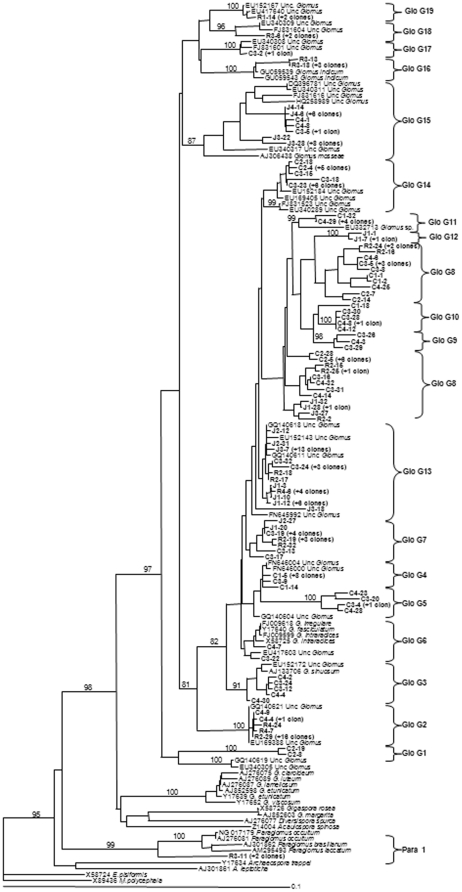
Figure 1. Neighbour-Joining (NJ) phylogenetic tree showing AM fungal sequences isolated from of native vegetation soil and Ricinus communis and Jatropa curcas soil and reference sequences from GeneBank.
All bootstrap values >85% are shown (1000 replicates). Numbers above branches indicate the bootstrap values of the NJ analysis; numbers below branches indicate the bootstrap values of the maximum likelihood analysis. Sequences obtained in the present study are shown in bold type. They are labelled with the database accession number, the soil from the host plant from which they were obtained (C = native vegetation soil, R = R. communis soil and J = J. curcas soil) and the clone identity number. Group identifiers (for example Glo G1) are AM fungal sequences types found in our study. Endogone pisiformis and Mortierella polycephala were used as out-groups.