Abstract
Accurate interpretation of recruitment rate measurements of microscale particles, such as cells and microbeads, to biofunctional surfaces is difficult because factors such as uneven ligand distributions, particle collisions, variable particle fluxes, and molecular-scale surface separation distances obfuscate the ability to link the observed particle behavior with the governing nanoscale biophysics. We report the development of a hydrodynamically conditioned micropattern catch strip assay to measure microparticle recruitment kinetics. The assay exploited patterning within microfluidic channels and the mechanostability of selectin bonds to create reaction geometries that confined a microbead flux to within 200 nm of the surface under flow conditions. Systematic control of capillary action enabled the creation of homogeneous or gradient ligand distributions. The method enabled the measurement of particle recruitment rates (keff, s−1) that were primarily determined by the interaction of the biomolecular pair being investigated. The method is therefore well suited for relative measurements of delivery vehicle and cellular recruitment potential as governed by surface-bound molecules.
The scientific and clinical importance of understanding the recruitment of leukocytes, stem cells, metastatic cancer cells, medical imaging contrast agents, and drug delivery vehicles to tissues has led to the development of a number of assays to analyze each stage in the process. A frequently exploited technology is the in vitro flow chamber assay. Studies of adhesion employing flow chambers require culturing cells1 or immobilizing ligands on a target surface,2,3 flowing a suspension of cells over the surface and observing interactions. Despite their inherent complexity, flow chamber assays have led to quantitative insights into some of the biophysical factors important for cell adhesion, especially in the context of cellular dissociation rates.4
Although the flow chamber assay has been used successfully to deduce the apparent bond lifetimes of mechanically stressed cell adhesion molecules, several factors have made the determination of a molecularly mediated capture parameter intrinsic to the microparticle more difficult. For one, the flux of microparticles sufficiently close to the surface to engage in adhesive interactions changes with the wall shear rate and position in the flow chamber.5,6 In addition, factors such as the hydrodynamic effects of bound microparticles on the flow field7 or collisions between microparticles8 may have an impact on accumulation on the surface. Finally, depending on the immobilization technique employed, the adhesive ligand may not be evenly distributed on the surface.9,10
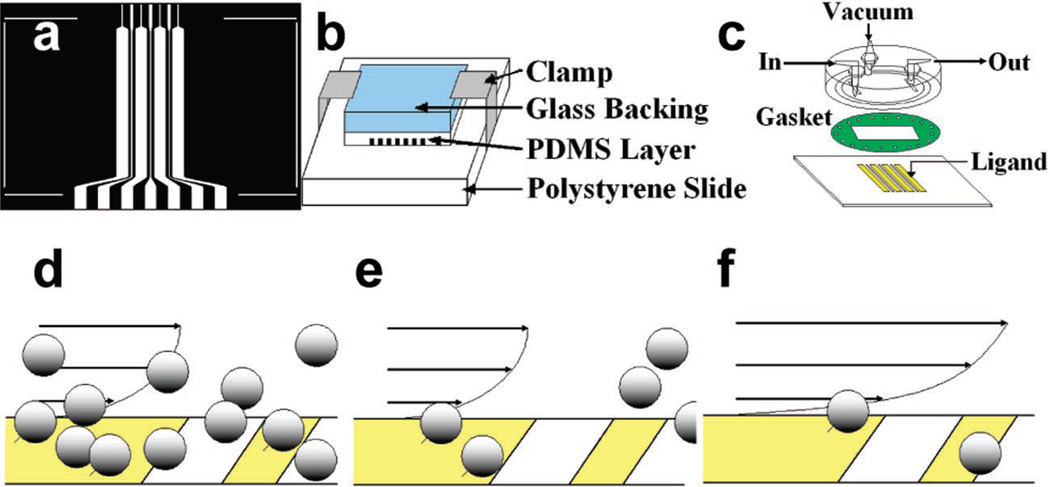
We developed an assay that geometrically controlled the presentation of adhesive ligand to establish a defined microparticle population flux for measurement. Wide accumulation strips (Figure 1a) of P-selectin glycoprotein ligand-1 (PSGL-1) were used to accumulate particles presenting recombinant P-selectin IgG chimera (P-rIgG). Once a sufficient quantity of particles accumulated (Figure 1d), unbound particles were rinsed away (Figure 1e), and the flow was increased to yield the desired wall shear rate. The probability that particles which dissociated from the accumulation strip were recruited to the downstream catch strip was measured (Figure 1f). Analysis of the recapture probability enabled the determination of an effective recruitment rate for the particle.
Figure 1.
Design, preparation, and implementation of the catch strip assay. (a) Pattern for photolithography. The pattern was 2 cm wide by 1 cm tall on the photomask. The narrowing of the channel where fluid entered the wide accumulation strips restricted the flow and helped to minimize the entrainment of air pockets in the fluid as the solution was drawn in. (b) Immobilization of the capillary mask to the slide for patterning. A glass backing helped to distribute the pressure. (c) A parallel plate flow chamber was immobilized to the slide with negative pressure. (d) After the flow chamber was mounted on the patterned surface, beads were flowed through so they could adhere to the accumulation strips. (e) Once a sufficient quantity of beads had accumulated, bead-free flow buffer was used to wash away unbound beads. (f) Flow was adjusted to yield the desired wall shear rate. Beads were observed to detach from the accumulation strip and jump the gap. The number of detaching microbeads that were recruited to the catch strip and the number that passed without a recruitment event were counted, thereby establishing a near-wall particle flux independent of sedimentation effects.
The assay was designed to address complications that obfuscate the deduction of kinetic parameters from flow chamber measurements. The elimination of a free air–water interface within the microchannels should eliminate fluid convection during the adsorption process and result in a surface with a more uniform protein coating.9–11 The assay configuration also mitigated the hydrodynamic effect of prebound microparticles on the recruitment rate by enabling the exclusion of particles that closely approached others from the measured population. An illustrative movie is available online (Supporting Information).
EXPERIMENTAL SECTION
Design and Fabrication of Capillary Masks
A rapid prototyping method similar to that described by Duffy et al. was implemented.12 Photomask patterns were designed in Illustrator (Adobe Systems Inc., San Jose, CA) and printed on a transparency with a 3600 dpi laser printer. Patterns of SU-8 2035 (MicroChem, Newton, MA) were created on a silicon wafer with photolithography. Poly(dimethylsiloxane) (PDMS) capillary masks were created by vapor deposition of (1H,1H,2H,2H)-perfluorooctyltrichlorosilane (Lancaster Synthesis, Pelham, NH) onto the masters, coating with PDMS (Sylgard 184, Dow Chemical Co., Midland, MI), degassing under a vacuum for several hours, adding a glass backing, and curing overnight at 60 °C. Channels 35 µm tall were found to have satisfactory capillary action. PDMS layers between 220 µm and 1 mm thick were able form a good seal with the polystyrene substrate but were not so thick that the microchannels collapsed.13
Preparation of Surfaces
The capillary mask was treated for 4–16 s with an LT III oxygen plasma gun (Lectro Engineering Co., St. Louis, MO) at 70% power to increase the hydrophilic characteristic of the PDMS.12 The capillary mask was reversibly secured to slides cut from untreated polystyrene culture dishes (Fisher Scientific, Suwanee, GA) with clamps (Figure 1b). A thick piece of glass clamped to the bottom of the polystyrene slide helped to ensure rigidity and better distribute the pressure (not shown in Figure 1b). We developed separate methods for adsorbing the proteins onto the slides to create either a homogeneous ligand distribution or a gradient. For both methods, protein solution (30 µL) was placed at the capillary mask’s entrance, which corresponded to the top of the features in Figure 1a. Capillary action drew the solution through the microchannels. Allowing the protein to adsorb without drawing additional fluid volume through the channels resulted in a gradient. If a homogeneous ligand distribution was desired, the remaining protein solution was drawn through the channels by wicking into bibulous paper, and an additional 30 µL was drawn through in the reverse direction. Incubation was performed for 3 h at room temperature. Wicking 10 µL of 0.2-µm filtered 1% casein (Pierce, Rockford, IL) immediately prior to mask removal minimized protein deposition in the gap regions between patterned ligand. Tetramethylrhodamine-labeled bovine serum albumin (tmr-BSA; Invitrogen, Eugene, OR) was used when a fluorescent marker was needed. PSGL-1 was isolated as described previously.2 Slides were blocked overnight at 4 °C with 1% Tween 20 (EMD Biosciences, La Jolla, CA) in Hank’s balanced salt solution (HBSS; Invitrogen).
Flow Experiment Preparation
Polystyrene microbeads (Polysciences, Inc., Warrington, PA) were incubated with P-rIgG (R&D Systems, Minneapolis, MN) for 3 h at room temperature. The solution P-rIgG concentration was adjusted to 5.8 × 10−10 µg/µm2 based on the bead surface area in a volume of 0.5–1.5 mL during adsorption. The microbeads were blocked overnight at 4 °C with 1% Tween 20. Flow experiments were performed by attaching a vacuum-immobilized flow chamber (GlycoTech, Gaithersburg, MD) to the functionalized surface as described elsewhere.27 Optical verification of the height of the gap in the mounted flow chamber agreed with the gasket design dimensions to within a few percent. Flow buffers were 0.2 µm filtered and made with CaCl2 diluted to 2 mM in HBSS. Inclusion of 1% Tween 20 by volume reduced microbead aggregation in the buffer. For experiments investigating effects of the microparticle receptor density on the recapture probability, microbead site densities were measured with fluorescein-labeled monoclonal antibody G1 (Ancell Corp., Bayport, MN) by comparison with Quantum Simply Cellular Anti-Mouse IgG Antigen Binding Capacity standards (Bang’s Laboratories, Inc., Fishers, IN) on a FACSCalibur laser benchtop cytometer (Becton Dickinson, Franklin Lakes, NJ) and background corrected by comparison with negative control microbeads.14
Functional Characterization of Ligand Distribution
Selectins immobilized on microbeads are capable of mediating stochastic, reversible molecular binding events with immobilized ligands; the process is called rolling due to the resulting tumbling motion. It has been shown that the rolling velocity of microparticles is a sensitive reporter of relative site density,15 so analyzing average rolling velocities mediated by the P-rIgG/PSGL-1 pair enabled us to investigate local variations in adsorbed protein density.
High Spatiotemporal Resolution Characterization of Microbead Detachment
Microbeads detaching from the accumulation strip were tracked in order to determine whether the gap size employed was sufficient to allow for acceleration to a steady velocity. A high-speed Fastcam PCI-R2 (Photron, San Diego, CA) was used to observe 6-µm beads detaching from the ligand-presenting area with 40× magnification at wall shear rates of 50 and 100 s−1. These experiments were performed on an active vibration isolation table (Micro-g 61-510-01, Technical Manufacturing Corp., Peabody, MA). Details on the tracking methodology are available in the Supporting Information. To investigate whether there was PDMS left behind that had an observable effect on the microparticle motion, a functional edge was also created on an unpatterned slide by adsorbing protein from a 30-µL droplet. A solution of 0.5 µg/mL PSGL-1 and 25 µg/mL tmr-BSA was used for both surfaces.
Determination of keff
Unlabeled human serum albumin (HSA; ZLB Bioplasma, Berne, Switzerland) and BSA (Sigma, St. Louis, MO) were used as diluent proteins when surfaces were prepared for the catch strip assay. A solution of 10 µg/mL HSA and 1 µg/mL PSGL-1 was used for high ligand coating concentrations, and for a low coating concentration, a solution of 50 µg/mL BSA and 0.25 µg/mL PSGL-1 was used. Measurements were performed with 10-µm beads on the high-density strips and with 6-µm beads on the lower density strips. Videos were acquired using a 20× objective and an analog videocamera (CCD72S, Dage-MTI, Michigan City, IN) and recorded to tape with a VCR.
Beads were observed as they detached from the accumulation strip. It was noted whether a microbead was already present within one particle diameter of the projected flight path of the microbead over the catch strip. If a microbead was already adhered within the zone of three diameter widths, the detaching microbead was not included in the analysis, effectively reducing one large source of flow-field disturbances. Recruitment was defined as an event where a bead was observed to occupy the same position for at least three consecutive frames (moving less than 1 pixel for 0.1 s) to facilitate manual analysis and ensure the particle effectively paused on the surface. Small variations in the required minimum number of stationary frames had little effect on the measured recruitment probability (e.g., 2 frames instead of 3). At least 20 microbeads were observed for each of the features on the slide for each experimental condition, but more observations were made when the flux was high. The average hydrodynamic velocity of 10 beads at each condition was measured after they detached from the accumulation strip and were passing over the gap region.
RESULTS AND DISCUSSION
Verification of Pattern
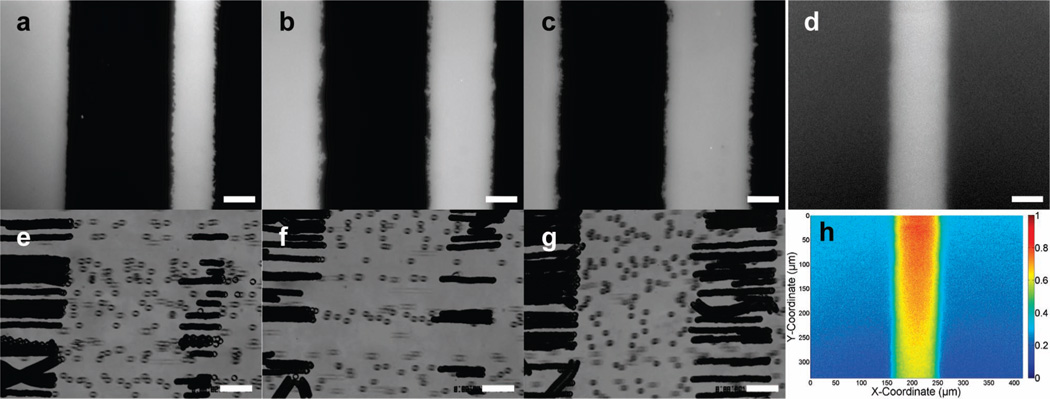
Microfluidic patterning of both fluorescent proteins and functional ligands was performed to assess the quality and dimensions of the patterns. Qualitatively, the patterns of adsorbed, fluorescent proteins (Figure 2a–c) agreed well with the strobelike functional maps created from the catch assay video (Figure 2e–g). The increase in velocity as a previously interacting bead traversed the gap was apparent from the greater spacing between successive bead images. The strobe images of individual beads detaching from the accumulation strip indicated a boundary of adhesive protein that functionally defined the gap. All optical and functional measures suggested a gap of 175 ± 10 µm for each of the slide areas examined. Optical and functional measures of the three catch strip sizes agreed within 5 µm and they were approximately 60, 95, and 130 µm wide.
Figure 2.
Visual and functional verification of the pattern. The scale bar in all images is 50 µm. (a–c) The patterned areas were verified by imaging patterned tmr-BSA. (e–g) Functional maps of 10-µm beads on the patterned slides at a wall shear rate of 67 s−1, 330 ms between overlays. Maps were acquired in experiments separate from the fluorescent images shown in the row above. The edge forming the end of the accumulation strip had an angle of 83° from the horizontal in (e, f) and 87° in (g). There was a ~4° discrepancy in camera angle between (g) and (e, f). Inspection of the flight paths of individual microbeads suggested a 5° alignment error of the flow chamber with the pattern. (d) A 5 µg/mL solution of tmr-BSA was drawn through the capillary mask with capillary action, and no additional volume exchange steps were applied. The background-corrected intensity is shown. (h) Pixel intensities from (d) were normalized and color-coded.
Wicking steps were used to introduce additional protein solute and control the surface distribution of the tmr-BSA. Drawing a solution of fluorescently labeled protein through the microfluidic mask in both directions resulted in a coating of consistent fluorescent intensity. Omission of the additional fluid exchange and utilization of solutions with a low total protein concentration resulted in a decreasing intensity along the length (Figure 2d). Normalization revealed a stronger intensity at portions of the feature closer to the end where fluid entered the channels and also near the center of the catch strip (Figure 2h). The decrease in adsorbed protein cumulatively resulted in striking differences in fluorescent intensity when comparing fields of view from different positions along the length of the strip. On the scale of the catch strip length (1 cm), a gradient of fluorescent protein could be created with concentrations up to 25 µg/mL.
Functional Characterization of Ligand Distribution
We oriented the flow field along the length of the catch strips and measured the average rolling velocities of P-rIgG-coated microbeads at different positions along the micropatterned strip to obtain a functional measure of local ligand concentration. We confirmed the ability to generate gradients and found that interassay and intra-assay variation were not statistically significant when high ligand-coating concentrations were employed, but variability was apparent when the functional ligand was very dilute (e.g., 0.05 µg/mL PSGL-1 stock solution and 25 µg/mL tmr-BSA). A more detailed presentation of these results can be found in the Supporting Information.
The ability to create micropatterned gradients suggested the method might be a basis for an inexpensive way to construct a diagnostic tool to functionally measure circulating populations of leukocytes, platelets, and even other tissue and stem cell populations according to their ability to exhibit distinct adhesive, morphological, or migrational16 responses. A functional cytometry approach to characterizing cell populations could also exploit combinations of chemotactic factor gradients and homogeneous adhesion molecule coatings presented on strips. For example, the ratio of leukocytes in a blood sample that migrated and aligned on a strip of SDF-1α and LFA-1 to those that migrated and aligned on a strip of SLC and ICAM-1 would give a ratio of effector to naïve T-cells, which could characterize the progression of an immune response during an infection.
Hydrodynamic Characterization of Microbead Detachment
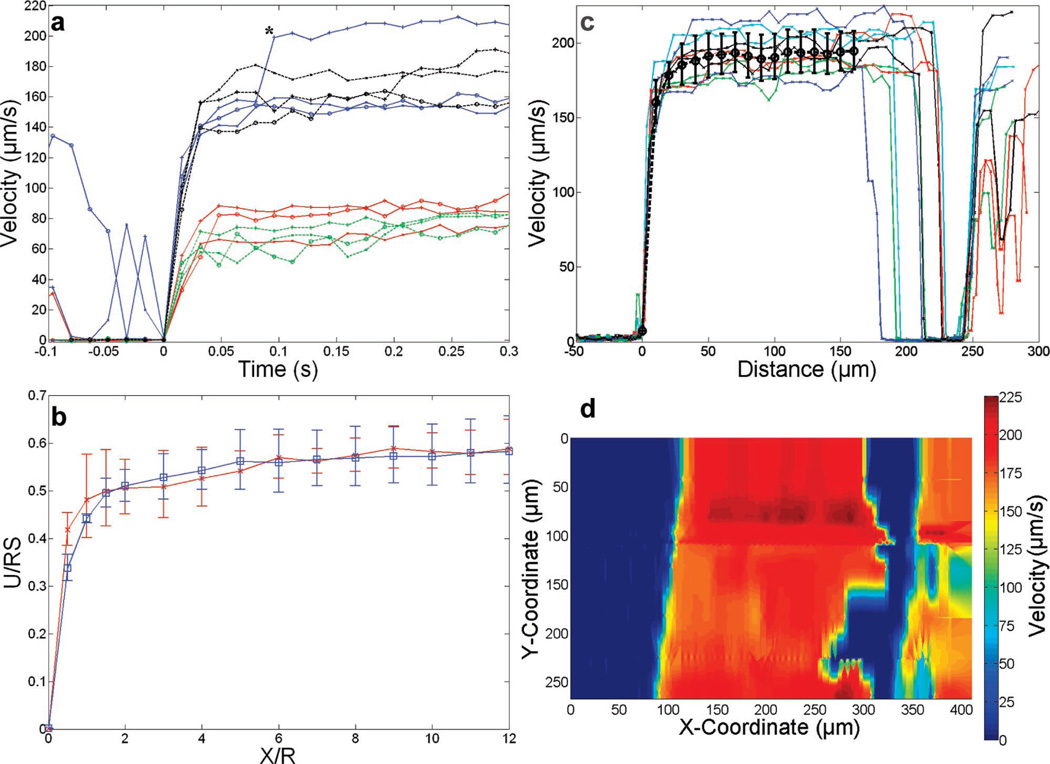
In order to validate the selection of gap size, 6-µm microbeads were computer-tracked with high spatiotemporal resolution as they detached from an edge of adsorbed protein. Experiments were performed at wall shear rates of 50 and 100 s−1, relevant for selectin-mediated rolling (Figure 3a). The velocity increased rapidly with distance from the detachment point initially, and the acceleration was essentially complete within one microbead diameter, 6 µm, for all of the beads observed. These data were averaged and nondimensionalized according to the parameters employed by Goldman et al. (Figure 3b).17 The dependence of the nondimensional velocity on nondimensional distance from the microbead detachment point was found to be very similar at wall shear rates of 50 and 100 s−1 and also suggested there might be gradual increase in the mean velocity up to six diameters from the detachment point. The results with the 6-µm beads were confirmed by tracking 10-µm beads in the lower resolution catch assay, which also attained steady velocity by about six microbead diameters (60 µm, Figure 3c).
Figure 3.
Investigation of microbead acceleration in the gap region lacking PSGL-1. (a) Microbead x-velocity as a function of time after detachment. Microbeads with a diameter of 6 µm were observed to detach from an accumulation strip at a wall shear rate of 50 (red tracings) and 100 s−1 (blue tracings). Microbeads detaching from an edge created by adsorption from a droplet at a wall shear rate of 50 (green dashed tracing) and 100 s−1 (black dashed tracing) behaved similarly to microbeads passing over the patterned gap, suggesting there were no PDMS particles left on the surface. *A rapid increase in velocity of the blue tracing indicated by plus symbols at 0.08 s after detachment occurred as an out-of-focus bead passed over the tracked bead. (b) Plots of the nondimensional geometric average velocity (left axis: velocity normalized by the microbead radius and wall shear rate) as a function of the nondimensional distance (bottom axis: distance normalized by the microparticle radius) from the detachment point at 50 (red) and 100 s−1 (blue). Error bars indicate the geometric standard deviation. (c) Tracking results for 10 of the 10-µm beads used to create the functional map in (d) at a wall shear rate of 67 s−1. The mean is shown in heavy black circles. (d) Heat map generated by spatial interpolation of microbead velocities.
A review of the video used to generate the high spatiotemporal resolution measurements of microbead velocity suggested hydrodynamic interactions between flowing microbeads could influence their velocity (Figure 3a, microbead at 100 s−1 with sudden increase in velocity). The velocity of a spherical object in shear flow is dependent on the separation distance from the plane it is traveling over.17,18 Hydrodynamic interactions with other unbound particles might affect the separation distance of the microbead from the surface, altering the average reactive contact area of the microbead and the binding probability. Measurements of recruitment events in assays that do not control the interacting population may vary as a function of both the microparticle concentration suspended in the buffer and the shear rate due to a changing frequency of interactions between microparticles.
Figure 3c presents x-velocity data from 10 of the microbeads as a function of position on the slide in the catch strip assay. An inverse distance-weighted interpolation method was used to obtain velocities in the coordinate system of the slide (Figure 3d). The choppy appearance at the leading edge of the catch strip, where the nearest data point neighbors to the grid points could switch between recruited and noninteracting microbeads, was a result of the interpolation technique. The plot demonstrated the stochastic nature of microbead recruitment, which was also apparent from the spatially staggered capture events in Figure 3c. Deceleration due to microbead capture occurred over shorter distances than acceleration due to detachment, which was apparent from the steeper color gradient in the heat map to the left relative to the right of blue areas. Figure 3d revealed some areas in the gap downstream of the catch strip that had functional molecules, but they were not problematic for the catch assay since the strip edges were still functionally intact.
Determination of keff
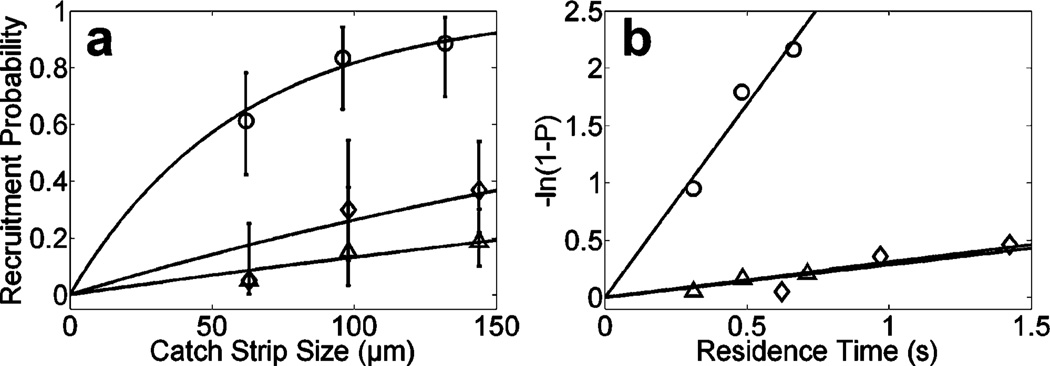
Microbead recruitment was measured in the catch assay under several different shear, particle size, and ligand-coating conditions (Figure 4a). A Poisson probability distribution function and an exponential cumulative distribution function describe events that have a constant probability of an occurrence per unit time. The distance until a recruitment event occurs should be governed by a Poisson distribution for identical particles under identical conditions.19, 20 A more detailed analysis is presented in the Supporting Information. A linear transformation for Poisson-distributed data was employed on the recruitment probability data, and the slope of the least-squares line with zero intercept was used to measure the effective recruitment rate of the microbeads (keff, Table 1). The linearity of the transformation suggested a Poisson model was approximately valid at both low and high adhesion probabilities and that the assay’s dynamic range was sufficient (Figure 4b). The agreement of the parameter calculated from the 6-µm bead recruitment data at 50 and 100 s−1 (insignificant p-value between the 6-µm diameter beads, Table 1) suggested the recruitment probability depended primarily on the contact time between the bead and surface. Upon normalization by the PSGL-1 concentration of the coating solution and reactive contact area (which scales with the microparticle radius), differences at high and low ligand densities were not significant, suggesting keff was a titratable indicator of molecular binding efficiency (Table 1, far right column).
Figure 4.
Determination of keff. (a) Microbead adhesion probability as a function of catch strip size. Curved lines: exponential cumulative distribution functions assuming the keff measured in (b). Error bars: 95% confidence interval. ○, 10-µm beads at 67 s−1 with concentrated patterned ligand; Δ, 6-µm beads at 100 s−1 with dilute patterned ligand; ◇, 6-µm beads at 50 s−1 and dilute patterned ligand. (b) Calculation of keff. The residence time was calculated by dividing the catch strip size by the average velocity. ○, keff = 3.38 s−1, R2 = 0.94; Δ, keff = 0.31 s−1, R2 = 0.84; ◇, keff = 0.29 s−1, R2 = 0.74.
Table 1.
Average Surface Separation Distances and Comparison of Recruitment Rates in the Catch Assay
| nominal diameter, µm |
mean radius, µm |
shear rate, s−1 |
average velocity, µm/s |
calculated separation, nm |
kBrownian, s−1 |
keff, s−1 |
reactive cap, nm |
keff/ [PSGL-1]*R mL/(µg µm s) |
|---|---|---|---|---|---|---|---|---|
| 10 | 4.7 | 67 | 199 | 142 | 0.41 | 3.4*,** | 0.3 | 0.72*** |
| 6 | 2.9 | 100 | 202 | 141 | 1.10 | 0.29* | 0.2 | 0.39*** |
| 6 | 2.9 | 50 | 101 | 141 | 1.10 | 0.31** | 0.2 | 0.42*** |
P = 0.008;
P = 0.007;
no significant differences (P > 0.01).
The design dimensions of the gap and catch strips required balancing photomask resolution limitations, hydrodynamics of particles in shear flow and the observation window size constrained by the laboratory microscope. The minimum feature size that could be achieved with the photomask manufacturing method employed was ~50 µm. Raggedness in the feature edges should not have an impact on the assay since the fluorescent protrusions (Figure 2a) were ~5 µm, and the characteristic binding distances, U/keff, were 59, 670, and 330 µm for the three conditions investigated. Given that the nondimensionalized velocity curves were very similar at 50 and 100 s−1 and the Reynolds number was much smaller than the transitional range to turbulent flow, the same gap dimension could likely be employed to probe a much wider range of physiologic shear than demonstrated here.
Theoretical investigation has demonstrated convection-enhanced reaction rates for rolling microparticles,20,21 and experimental investigations lacking a hydrodynamic conditioning step have indicated a shear-dependent increase in the rate of initial molecular tether formation for L-selectin bearing microbeads.21,22 The experimental observations demonstrate the importance of future measurements with the catch strip assay to investigate the effects of convection and force on the binding of all three selectin family members and performing the assay over a broad range of shear rates to better elucidate the important biophysical contributions to molecular tether formation, especially with respect to leukocyte recruitment.
A patterning approach employing multiple functional strips23,24 could facilitate a high-throughput targeting investigation with the catch strip assay. Microbeads could be functionalized with P-selectin, to facilitate hydrodynamic confinement of the microparticle, and the molecular target of interest. An accumulation strip would present PSGL-1, and the candidate targeting proteins would be presented on downstream catch strips. The assay could also be made combinatorial by observing differentially pigmented microbeads presenting different molecular targets with color video acquisition. One way to achieve more consistent ligand densities with lower coating concentrations for targeting screens would be to use syringe-driven flow to fill individual microchannels and perform volume exchanges.
Evaluation of Surface Separation Under Shear
The maximal PSGL-1-P-selectin bond length has been estimated as ~90 nm from electron micrographs.25,26 Several metrics were used to determine whether the observed microbead recruitment events were indicative of the reaction kinetics or if the approach of the particles to within a suitable binding distance from the surface was diffusion limited (Table 1). The estimated average separation distance calculated from the hydrodynamic velocity was ~140 nm and agreed very well across the shear rates and bead sizes investigated. A 20-µm-long AFM scan line of the polystyrene material, available in the Supporting Information, had a peak roughness of 27 nm. It is possible the polystyrene microbeads had a similar roughness, so the data strongly suggested surface roughness helped bring the molecular binding partners within binding distance despite the large average separation distance calculated from hydrodynamics. The vertical motion of a microbead close to the wall is highly damped. The spatially weighted diffusional coefficient between 140 and 90 nm for 10- and 6-µm beads was calculated to be 43 and 27 times smaller than for microbeads far from the surface, respectively.27 The observation that the 10-µm beads were recruited faster than this theoretical rate limitation (kBrownian, Table 1) supported the existence of a role for surface roughness, as did the observation that the 10-µm beads were recruited more quickly than the 6-µm microbeads. An analysis of published protein adsorption efficiencies and two-dimensional reaction rate indicated that only 1 nm of the microbead cap was within binding distance (reactive cap, Table 1, calculation described in the Supporting Information), which suggested the microbeads were at the extreme of their binding distance. Stochastic excursions of a few nanometers could facilitate molecular tethering without being rate limiting.
At the estimated average separation distance, successful binding events would require P-selectin and PSGL-1 to be fully extended. Interestingly, studies employing receptor–ligand pairs attached to surfaces by chains of poly(ethylene glycol) (PEG) found maximal chain extension was a rare event that limited how quickly surfaces could bind.28 Full molecular extension might be a biologically selected feature that enhances molecular recognition under flow. Indeed, it has been noted that P-selectin and PSGL-1 are both rigid molecules that extend far from the membrane they are tethered to due to limited interdomain flexibility and a high degree of glycosylation, respectively.25,26 The direct adsorption methods employed in this assay did not ensure the orientation of the P-rIgG or the PSGL-1, although the hydrophobic transmembrane domain in the PSGL-1 molecule used may serve as a preferential adsorption point. In the future, orientation could be achieved by using adsorbed antibodies to ensure attachment of the ligand to the surface at a specific point,29 incorporating transmembrane ligands into liposomes to form lipid bilayers on a glass surface,23 attachment of biotinylated ligand molecules to a streptavidin-functionalized surface through a PEG linker,30 or using a heterobifunctional PEG cross-linker to covalently attach the ligand to a functionalized surface.
Total internal reflection fluorescent microscopy (TIRFM), might yield further insight into whether the reactive end groups of macromolecules effectively sample the entire vertical distance from the membrane allowed by the tether structure and enable an absolute measure of reaction kinetics.31,32 Recent studies implementing TIRFM have revealed that particles could be up to 100 nm closer to the wall than would be expected from velocity measurements with sufficiently smooth surfaces, and direct measurement of this separation would be preferable to indirect measurements calculated from velocity or Brownian motion data.33,34 Previous studies that have yielded estimates of two-dimensional reaction rates for the selectins have involved pushing the opposing surfaces closely together under static conditions,29,35 which removes effects of force, molecular freedom,36 and contact time and further limits the assay to a low-throughput mode.25,26 The catch strip assay has an inherent advantage for measuring 2D kinetics governing recruitment since the reaction configuration better resembles physiologic conditions.
Assay Utility
The density of reactive selectin binding domains on the microparticles was varied. We report stock dilutions rather than protein concentration because more impurities were present in the second PSGL-1 preparation than the previous; however, this would not affect the applicability of the result. The recruitment probability scaled well with the P-selectin site density, further validating the assay as a valid means of obtaining a relative measure of microparticle recruitment kinetics (Table 2).
Table 2.
Effect of Microparticle Site Density on Relative Recruitment Kineticsa
| slide |
mPsel, sites/µm2 |
SD, sites/µm2 |
keff, s−1 |
SD, s−1 |
keff/mPsel. µm2/s |
|---|---|---|---|---|---|
| 1 | 15 | 0.9 | 0.61 | 0.36 | 0.0400 |
| 37 | 11 | 1.54 | 0.54 | 0.0420 | |
| 2 | 18 | 16 | 0.76 | 0.05 | 0.0420 |
| 47 | 14 | 1.46 | 0.30 | 0.0310 |
Slides were patterned with a 1:10 dilution of stock PSGL-1 solution and 10 µg/mL HSA. The reported site is the estimated number of P-selectin binding pockets (lectin domains). Experiments were performed with 6-µm particles at a wall shear rate of 100 s−1.
CONCLUSIONS
Relative to measurements of adhesive bond lifetime, few normalized measurements of capture and recruitment to surfaces have been reported. Recruitment remains an important parameter to quantify in order to elucidate the dominant molecular interactions in leukocyte homing to inflamed endothelium and tissue, facilitate the design of microscale delivery vectors, and develop models of fast, two-dimensional reaction kinetics.37 The catch strip assay created a hydrodynamically conditioned population of microparticles and enabled a quantitative measurement of recruitment rates that were dependent upon biomolecular recognition events. The measured recruitment rate, keff, enabled the comparison of microparticle adhesive potential over a range of recruitment conditions. In addition to the relative measurements presented here, an extension of the assay will facilitate an analysis of absolute 2D on-rates when coupled with a technique to assess instantaneous separation, such as TIRFM, and may form a basis for functional cytometry applications.
Supplementary Material
ACKNOWLEDGMENT
The authors gratefully acknowledge financial support from the Biotechnology Training Program (NIGMS 5T32GM008715) and a Biotechnology Research Partnership award (NIHEB002185-05). We thank Dr. Brian P. Helmke of the University of Virginia and Dr. David W. Schmidtke of the University of Oklahoma for their guidance on micropatterning strategies as well as Bryan R. Smith of the University of Virginia for technical discussions and excellent material support.
Footnotes
SUPPORTING INFORMATION AVAILABLE
A movie demonstration of the assay, functional analysis of ligand distribution, analysis of applicability of the Poisson distribution, AFM results, and additional details on the patterning, tracking, and statistical methodology. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lawrence MB, McIntire LV, Eskin SG. Blood. 1987;70:1284–1290. [PubMed] [Google Scholar]
- 2.Park EY, Smith MJ, Stropp ES, Snapp KR, DiVietro JA, Walker WF, Schmidtke DW, Diamond SL, Lawrence MB. Biophys. J. 2002;82:1835–1847. doi: 10.1016/S0006-3495(02)75534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence MB, Springer TA. Cell. 1991;65:859–873. doi: 10.1016/0092-8674(91)90393-d. [DOI] [PubMed] [Google Scholar]
- 4.Smith MJ, Berg EL, Lawrence MB. Biophys. J. 1999;77:3371–3383. doi: 10.1016/S0006-3495(99)77169-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munn LL, Melder RJ, Jain RK. Biophys. J. 1994;67:889–895. doi: 10.1016/S0006-3495(94)80550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Neelamegham S. Biophys. J. 2002;83:1934–1952. doi: 10.1016/S0006-3495(02)73956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King MR, Hammer DA. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14919–14924. doi: 10.1073/pnas.261272498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadash KE, Lawrence MB, Diamond SL. Biophys. J. 2004;86:4030–4039. doi: 10.1529/biophysj.103.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deegan RD. Phys. Rev. E: Stat. Phys. Plasmas, Fluids, Relat. Interdiscip Top. 2000;61:475–485. doi: 10.1103/physreve.61.475. [DOI] [PubMed] [Google Scholar]
- 10.Hu H, Larson RG. Langmuir. 2005;21:3963–3971. doi: 10.1021/la047528s. [DOI] [PubMed] [Google Scholar]
- 11.Balagurunathan Y, Dougherty ER, Chen Y, Bittner ML, Trent JM. J. Biomed. Opt. 2002;7:507–523. doi: 10.1117/1.1486246. [DOI] [PubMed] [Google Scholar]
- 12.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 13.James CD, Davis RC, Kam L, Craighead HG, Isaacson M, Turner qJN, Shain W. Langmuir. 1998;14:741–744. [Google Scholar]
- 14.Kerker M, Van Dilla MA, Brunsting A, Kratohvil JP, Hsu P, Wang DS, Gray JW, Langlois RG. Cytometry. 1982;3:71–78. doi: 10.1002/cyto.990030202. [DOI] [PubMed] [Google Scholar]
- 15.Brunk DK, Hammer DA. Biophys. J. 1997;72:2820–2833. doi: 10.1016/S0006-3495(97)78924-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Jeon N, Baskaran H, Dertinger SK, Whitesides GM, Van de Water L, Toner M. Nat. Biotechnol. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 17.Goldman AJ, Cox RG, Brenner H. Chem. Eng. Sci. 1967;22:653–660. [Google Scholar]
- 18.Pierres A, Benoliel AM, Zhu C, Bongrand P. Biophys. J. 2001;81:25–42. doi: 10.1016/S0006-3495(01)75677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tempelman LA, Hammer DA. Biophys. J. 1994;66:1231–1243. doi: 10.1016/S0006-3495(94)80907-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang KC, Hammer DA. Biophys. J. 1999;76:1280–1292. doi: 10.1016/S0006-3495(99)77291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paschall CD, Guilford WH, Lawrence MB. Biophys. J. doi: 10.1529/biophysj.106.098707. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yago T, Zarnitsyna VI, Klopocki AG, McEver RP, Zhu C. Biophys. J. 2007;92:330–342. doi: 10.1529/biophysj.106.090969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burridge KA, Figa MA, Wong JY. Langmuir. 2004;20:10252–10259. doi: 10.1021/la0489099. [DOI] [PubMed] [Google Scholar]
- 24.Nalayanda DD, Kalukanimuttam M, Schmidtke DW. Biomed. Microdevices. 2007;9:207–214. doi: 10.1007/s10544-006-9022-6. [DOI] [PubMed] [Google Scholar]
- 25.Li F, Erickson HP, James JA, Moore KL, Cummings RD, McEver RP. J. Biol. Chem. 1996;271:6342–6348. doi: 10.1074/jbc.271.11.6342. [DOI] [PubMed] [Google Scholar]
- 26.Ushiyama S, Laue TM, Moore KL, Erickson HP, McEver RPJ. Biol. Chem. 1993;268:15229–15237. [PubMed] [Google Scholar]
- 27.Brenner H. Chem. Eng. Sci. 1961;16:242–251. [Google Scholar]
- 28.Jeppesen C, Wong JY, Kuhl TL, Israelachvili JN, Mullah N, Zalipsky S, Marques CM. Science. 2001;293:465–468. doi: 10.1126/science.293.5529.465. [DOI] [PubMed] [Google Scholar]
- 29.Huang J, Chen J, Chesla SE, Yago T, Mehta P, McEver RP, Zhu C, Long M. J. Biol. Chem. 2004;279:44915–44923. doi: 10.1074/jbc.M407039200. [DOI] [PubMed] [Google Scholar]
- 30.Patel N, Padera R, Sanders GH, Cannizzaro SM, Davies MC, Langer R, Roberts CJ, Tendler SJ, Williams PM, Shakesheff KM. FASEB J. 1998;12:1447–1454. doi: 10.1096/fasebj.12.14.1447. [DOI] [PubMed] [Google Scholar]
- 31.Jin S, Huang P, Park J, Yoo JY, Breuer KS. Exp. Fluids. 2004;37:825–833. [Google Scholar]
- 32.Prieve DC. Adv. Colloid Interface Sci. 1999;82:93–125. [Google Scholar]
- 33.Huang P, Guasto JS, Breuer KSJ. Fluid Mech. 2006;566:447–464. [Google Scholar]
- 34.Huang P, Breuer KS. Phys. Fluids. 2007;19 028104. [Google Scholar]
- 35.Rinko LJ, Lawrence MB, Guilford WH. Biophys. J. 2004;86:544–554. doi: 10.1016/S0006-3495(04)74133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dustin ML, Ferguson LM, Chan PY, Springer TA, Golan DEJ. Cell Biol. 1996;132:465–474. doi: 10.1083/jcb.132.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dustin ML, Bromley SK, Davis MM, Zhu C. Annu. Rev. Cell Dev. Biol. 2001;17:133–157. doi: 10.1146/annurev.cellbio.17.1.133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.