Abstract
The present study analyzed the relationship between the genetic diversity of Plasmodium falciparum and parasitologic/entomologic indices in the Mount Cameroon region by using merozoite surface protein 1 as a genetic marker. Blood samples were collected from asymptomatic children from three altitude zones (high, intermediate, and low). Parasitologic and entomologic indices were determined by microscopy and landing catch mosquito collection/circumsporozoite protein–enzyme-linked immunosorbent assay, respectively. A total of 142 randomly selected P. falciparum-positive blood samples were genotyped by using a nested polymerase chain reaction–based technique. K-1 polymerase chain reaction products were also sequenced. As opposed to high altitude, the highest malaria prevalence (70.65%) and entomologic inoculation rate (2.43 infective/bites/night) were recorded at a low altitude site. Seven (18.91%), 22 (36.66%), and 19 (42.22%) samples from high, intermediate, and low altitudes, respectively, contained multiclonal infections. A new K-1 polymorphism was identified. This study shows a positive non-linear association between low/intermediate altitude (high malaria transmission) and an increase in P. falciparum merozoite surface protein 1 block 2 polymorphisms.
Introduction
Knowledge of the molecular epidemiology of parasites in an area is essential to understand the transmission of a disease such as malaria. Malaria is the most serious tropical parasitic disease and one of the major threats to humans worldwide. It causes 1–3 million deaths and more than 300 million clinical cases annually with more than 90% occurring in sub-Saharan Africa.1–3 Most of the cases are caused by Plasmodium falciparum, and children and pregnant women are the most affected group.3,4
Malaria control may be achieved by three complementary methods: chemotherapy, vaccination, and vector control. Nevertheless, the appearance and spread of resistance to antimalarial drugs and insecticides have become a primary concern for human health care.5–7 Although the need for a safe and effective malaria vaccine is more urgent than ever, antigenic and strain diversities of the parasite remain a major obstacle, which highlight mandatory appreciation of the diversity of the local parasite population before development of any immunologic interventions.8 This appreciation would be a step forward generating current and comprehensive information on the diversity in the genes that encode malaria vaccine candidates of P. falciparum and its implications on the epidemiology of malaria, immunity, and development of control measures.9 Plasmodium falciparum is genetically diverse at all levels of endemicity, which is not surprising because genetic diversity has been shown to be a function of transmission in a given area.10–12 The inherent variability of the P. falciparum is particularly prevalent in merozoite surface antigens or proteins (MSPs).13 These proteins provides multiple effective evasion and drug resistance mechanisms for the parasite. They also represent a major challenge for development of an effective malaria vaccine.
Merozoite surface protein 1 is the most commonly used genetic marker for the determination of the genetic diversity of the malaria parasite.14–16 Twenty-four major versions of the MSP-1 gene have been identified,17,18 and its coding sequence may be divided into 17 blocks among which 7 are variable blocks interspersed with conserved and semi-conserved regions. In some variable blocks, the variation is dimorphic and sequences may be grouped into one of the two allelic families (K-1 and MAD20). Block 2 represents an exception to dimorphism; it is the most polymorphic part of the gene having a third allelic family or variant RO33.19 Genetic diversity at the MSP-1 locus may be generated by exchanging blocks of sequences during sexual (meiotic) recombination and by putative strand-slippage events during asexual (mitotic) replication of the parasites, which lead to rearrangements of block 2 tripeptide repeats.17,20 High meiotic recombination rates within MSP-1 have been estimated for parasites in areas of intense malaria transmission in Africa, where most human infections consist of mixture of genetically distinct allelic variants.21 However, it should be noted that meiotic recombination is rare between allelic types, although it occurs in block 2 between MAD20 and RO33 and creates a fourth allele family known as MR.22 The effect of altitude and estimated rainfall on indices of malaria infection/transmission has been described in a study carried out in Tanzania in which P. falciparum prevalence was negatively associated with altitude. However, the relationship varied according to ecological setting, climate, vector species, topography, and host and parasite genetics.23
Malaria prevalence in the Mount Cameroon region is high (≥ 90%).24,25 It has previously been shown that malaria transmission is heterogeneous, and the highest transmission rate has been recorded at lower altitudes.26 The malaria transmission pattern in the Mount Cameroon region is greatly influenced by altitude, climatic, and bio-ecological variations. Serious environmental alterations that have taken place in the region that have been caused by rapid growth in populations, roads, and houses and agro-industrial activities of the Cameroon Development Cooperation have led to ecological changes, which together with other factors, such as rainfall, temperature, and humidity, affect the structure of the vector population and thus transmission of infection and probably the genetic diversity of the parasites circulating in the area.10 Although previous entomologic and parasitologic studies in this region have shown the influence of these changes on the heterogeneity of the malaria transmission pattern observed, none had been conducted to determine whether this variability necessarily translates into variation in the genetic diversity of P. falciparum in the region.
The purpose of this study was to investigate genetic variations in the P. falciparum MSP-1 block 2 in samples collected from asymptomatic school children in six localities in the Mount Cameroon region, including two regions at high altitudes where transmission is low, two regions at the intermediate altitudes where transmission has a mixed pattern, and two other regions at low altitudes where transmission is high. This genetic altitude-based characterization of P. falciparum infections in the Mount Cameroon area will provide new essential data on the parasite population diversity and implications for the epidemiology of malaria and development of appropriate control measures.
Materials and Methods
Study design.
The present study aimed to correlate genetic variation in the MSP-1 block 2 of P. falciparum with malaria endemicity in six locations of contrasting altitudes in the Mount Cameroon region. Geographic parameters of the study area such as temperature and area of residence of participants, and demographic data such as age and sex of participants were recorded during the sampling period. Climatic parameters were obtained from the Cameroon Development Cooperation weather station. Blood samples were collected during March–July 2006, which is the mild rainy season during which transmission of malaria is high in the region. Malaria prevalence, parasite density, and Anopheles biting and entomologic inoculation rates (EIRs) were determined.
A census was used for selection of participants. Patients eligible for this study were asymptomatic school children of both sexes 4–16 years of age. This particular age group was chosen for their relative settled way of life, thus reducing bias that can be caused by movements of populations and acquisition of different strains of parasite from other areas. Government schools were chosen because they were found in all localities, and most parents send their children to these schools because fees are moderate. Objectives and schedules of the study were explained to the parents or guardians of the eligible children, and those who agreed to allow their children to participate signed an informed consent form. Eligible children were enrolled in the study. This study was approved by the ethical review board of Kumba station and the Cameroonian Ministry of Public Health.
Study area.
The study was conducted in six localities that had contrasting altitudes, different malaria transmission profiles, and contrasting climatic/environmental features: (Bonakanda = 1,197 meters above sea level) and Likoko-Membea (800 meters above sea level) were considered high-altitude areas, Meanja (300 meters above sea level) and Mutengene (220 meters above sea level) were considered intermediate-altitude areas, and Debundscha (50 meters above sea level) and Tiko (10 meters above sea level) were considered low-altitude areas. Mount Cameroon in Buea, Cameroon, is the highest mountain in western Africa and is an active volcano that rises from the Atlantic Ocean at the Gulf of Guinea and has an altitude of 4,100 meters above sea level. The mountain is formed of a continuous pile of terraces from the base to the summit. At an altitude of 50 meters from the coast is a sedimentary plain that extends from Tiko to Debundscha. From Mutengene, the altitude of the terrain gradually increases to an altitude of 800–1,200 meters in Buea (Bonakanda).
Hydrologically, there are 20 streams in the study area that empty into the Atlantic Ocean. Two of these streams, Ombe and Onge, flow southeast and northwest, respectively. In this forested area of southern Cameroon, the equatorial climate is modified by the influence of the ocean and the mountain. At higher altitudes, the temperatures are lower than in other areas of the southern part of the country: mean values of the minimum temperatures are 20°C in December and 18°C in August, and mean values of the maximum temperature are 35°C in August and 30°C in March. Rainfall was also an important factor in this study. Debundscha, located at the western flanks of Mount Cameroon, has up to 11,000 mm of rainfall, making this locality the second wettest place in the world. The Mount Cameroon region has a long rainy season that starts in March and ends in November, and maximum rainfall occurs in August and September. The dry season starts in November and ends in February.
Approximately 1.3 million persons live near Mount Cameroon in the towns of Tiko, Limbe, Mutengene, Buea, and Muyuka. The population is composed of indigenous Bakweri, Creoles (from Liberia and Sierra Leone), and Nigerians and immigrants from other parts of Cameroon, especially the North West Province. Additional details on the features of villages studied in this work are shown in Table 1.
Table 1.
Climatic, environmental, and parasitologic characteristics of the study sites in the Mount Cameroon area
| Geographic coordinates | Site | Altitude (meters above sea level) | Mean temperature (°C) | Relative humidity (%) | Rainfall (mm) | Malaria prevalence (%) | Geometric mean parasite density/μL | |
|---|---|---|---|---|---|---|---|---|
| High altitude | 4°21′N, 9°27′E | Bonakanda | 1,197 | 19.5 | 86.4 | 2,400 | 12.33 | 522 |
| 4°14′N, 9°10′E | Likoko Membea | 800 | 22.5 | 88.8 | 2,654 | 16.82 | 496 | |
| Intermediate altitude | 4°07′N, 9°21′E | Meanja | 300 | 27.5 | 61.1 | 2,475 | 62.26 | 839 |
| 4°05′N, 9°18′E | Mutengene | 220 | 27.5 | 83.1 | 1,854 | 46.80 | 700 | |
| Low altitude | 4°10′N, 9°00′E | Debundscha | 50 | 27 | 89.6 | 10,000 | 42.66 | 650 |
| 4°07′N, 9°36′E | Tiko | 10 | 27.9 | 83.1 | 4,524 | 70.65 | 656 | |
Collection of blood samples and parasitologic examinations.
Using sterile needles and syringes, we collected 2 mL of venous blood from each participant into tubes containing EDTA according to routine clinical practice. Some blood was also collected onto grease-free slides for the preparation of thin and thick blood films and immediately spread to prevent clotting. In the laboratory, serum samples were separated from the blood cells by centrifugation at 2,000 rpm for 5 minutes, transferred into sterile Eppendorf (Hamburg, Germany) tubes, labeled accordingly, and stored at −20°C for future use. Blood cells were also stored at −20°C for DNA extraction. Thick and thin blood films were prepared and stained with 5% Giemsa according to the method described by Cheesbrough.27 Giemsa-stained films were examined by using the oil immersion objective (×100) of a light microscope for detection and identification of malaria parasites according to procedures of the World Health Organization.28 Slides were considered positive when asexual forms and/or gametocytes of P. falciparum were present in the blood film and negative after observing approximately 100 high-powered microscopic fields without seeing any parasites. For each positive slide, parasites were counted against 200 leukocytes and expressed as parasites per microliter of blood, assuming a leukocyte count of 8,000 leukocytes/μL of blood.27
Anopheles mosquito collection, human biting rates, and EIRs.
Mosquitoes were collected by the same work force at all stations (localities) during the entire study by standardized landing-catch collections. Mosquito collectors were given prophylactic treatment for malaria. All mosquitoes collected were morphologically identified by using the key of the Afro-Tropical Region and procedures for identification of Anopheles.29–31 Heads and thoraces of female Anopheles mosquitoes were obtained and tested for circumsporozoite proteins of P. falciparum by enzyme-linked immunosorbent assay as described.32 Human-biting rates (also known as aggressiveness of the species) (HBRs) were calculated directly from landing-catch collections as the average number of Anopheles bites experienced by a collector during an entire night of collection. The sporozoite rate was expressed as the proportion of Anopheles positive for sporozoites by an enzyme-linked immunosorbent assay for circumsporozoite protein of the total number tested. The EIR, expressed as the number of infective bites per person per night of collection, was calculated by multiplying the HBR by the sporozoite rate.33
Genomic DNA isolation from blood samples.
A total of 142 P. falciparum-positive blood samples were selected for genomic DNA extraction by using the QIAamp DNA blood midi Kit (QIAGEN, Crawley, United Kingdom) according to the manufacturer's instructions. In brief, 300 μL of whole blood from each sample was lysed and loaded onto a DNA affinity spin column. Genomic DNA bound to the membrane was purified by using two wash buffers to remove impurities. Genomic DNA was then was eluted with 100 μL of distilled water and stored at −20°C until use. The concentration and purity of isolated DNA samples were measured by using a spectrophotometer.
Amplification of MSP-1 allelic variants by polymerase chain reaction.
Amplification of MSP-1 block 2 was performed by using two rounds of nested polymerase chain reaction (PCR) according to the protocol described by Snounou and others.14 All oligonucleotide primers were obtained from Eurogentec (Seraing, Belgium). The polymorphic repetitive regions (block 2 of MSP-1) were amplified in a GeneAmp® PCR system 9700 thermocycler (Applied Biosystems, Foster City, CA) with the following cycling parameters: denaturation at 95°C for 5 minutes; followed by 35 cycles (first round) or 45 cycles (second round) at 95°C for 1 minute, 61°C (first round) or 58°C (second round) for 1 minute, and 72°C for 2 minutes; and final extension at 72°C for 5 minutes.
In the first-round PCR, sequences spanning blocks 1–5 of the MSP-1 gene were amplified with the outer sense primer M1-OF (5′-CTAGAAGCTTTAGAAGATGCAGTATTG-3′) and the outer antisense primer M1-OR (5′-CTTAAATAGTATTCTAATTCAAGTGGATCA-3′) and 1 μL of purified genomic DNA as template. Reactions were carried out in a final volume of 25 μL containing 50 mM KCl, 10 mM Tris-HCl, pH 9.0, 0.1% Triton X-100, 1.5 mM MgCl2, 0.25 mM of each of the four dNTPs, 0.4 μM of each specific primer, and 0.04 units/μL of Taq polymerase.
Three second-round nested reactions were subsequently performed to determine the presence of allelic variants from the MAD20, Kl, and RO33 types of the MSP-l block 2 by using each of the following pairs of nested specific primers: M1-MF 5′-AAATGAAGGAACAAGTGGAACAGCTGTTAC-3′ and M1-MR 5′-ATCTGAAGGATTTGTACGTCTTGAATTACC-3′; M1-KF 5′-AAATGAAGAAGAAATTACTACAAAAGGTGC-3′ and M1-KR 5′-GCTTGCATCAGCTGGAGGGCTTGCACCAGA-3′; M1-RF 5′-TAAAGGATGGAGCAAATACTCAAGTTGTTG-3′ and M1-RR 5′-CATCTGAAGGATTTGCAGCACCTGGAGATC-3, respectively. In all cases, 1 μL of a 1:70 dilution of amplification product from the first-round PCR was used in the three nested reactions. Positive controls (DNA from a parasite containing a specific MSP-1 block 2 allelic variant) and a negative control (without DNA) were included in each amplification.
The PCR products were subjected to electrophoresis on 2% agarose gels containing 0.1 mg/mL of ethidium bromide and visualized under UV light by using the Doc-print photographic system (Fisher Scientific Bioblock, Illkirch, France). Electrophoretic analysis of K-1-type PCR products showed bands of different sizes. To investigate the nature of the DNA fragments obtained by K-1 PCR-typing, these fragments were purified and cloned into the pGEM®-T Easy vector (Promega, Madison, WI) according to the manufacturer's recommendations and sequenced by GATC Biotech (Konstanz, Germany) by using the automated Sanger method (dideoxy-mediated chain termination).34 Homology searches in databases were performed using the basic local alignment search tool (BLAST) network service (National Center for Biotechnology Information, Bethesda, MD).
Data analysis.
Data was analyzed by using SPSS version 15 (SPSS Inc., Chicago, IL), and all tests were performed at a 5% level of significance. Different allelic variant frequencies were estimated by calculating the percentage of fragments assigned to one allelic variant type among the overall number of fragments detected for MSP-1 block 2. Gene types were defined according to the method of Da-Silveira and others18 as the unique combination of allelic types in variable blocks. The complexity of infection (mean number of lines or fragments) per sample was calculated for MSP-1 block 2 as the average number of distinct fragments (K-1, MAD20, and RO33) per PCR-positive sample.14 Parasite density was expressed as the geometric mean parasite density (GMPD) of P. falciparum per microliter of blood.
Significant changes in malaria prevalences and frequencies of allelic variants (K-1, MAD 20, and RO33) between localities and altitude zones were compared by using the chi-square test. The Kruskal-Wallis test was used to check for differences in the mean number of genotypes per sample between the three altitude zones. This test was used to account for the small sample size at high altitude and also because samples were not normally distributed. In addition, a non-parametric multiple comparison was performed by using the Dunn test to check for differences in the mean number of genotypes per sample between high and low altitudes, high and intermediate altitudes, and low and intermediate altitudes. This test was chosen because it is appropriate when the Kruskal-Wallis test fails to reject the null hypothesis and because it accounts for disparities in sample sizes. A Spearman correlation rank test was used to check the association between altitude and complexity of infection, malaria prevalence, GMPD, mosquito biting rate, and EIR.
Results
Parasitologic indices.
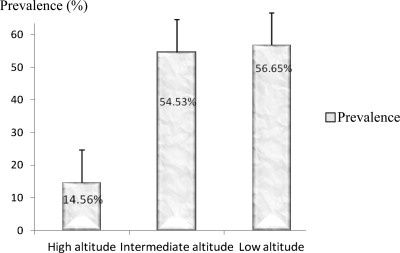
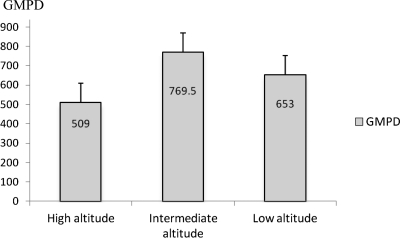
Blood samples were collected from 876 children. The overall prevalence of malaria parasites observed in the study population was 45.32% (397 of 876). The highest (70.65%) prevalence rate of malaria was recorded in Tiko (low altitude site), and the lowest (12.33%) was recorded in Bonakanda (high altitude site). An unexpected higher prevalence (62.26%) was obtained in Meanja (intermediate zone) than in Debundscha (42.66%), which is a low altitude site (Table 1). There was a significant difference in malaria prevalence between sites at different altitudes (P = 0.01), but the correlation between malaria prevalence and altitude was not significant (r = −0.771, P = 0.072). Globally, there was a trend of decreasing malaria prevalence with altitude (Figure 1). The GMPD of asexual stages of Plasmodium species was highly heterogeneous among different localities and intermediate zones had the highest value. A GMPD value of 839.42 parasites/μL of blood (range = 40–29,520 parasites/μL of blood) was recorded in Meanja (intermediate altitude site), and the lowest GMPD of 496.14 parasites/μL of blood (range = 120–4,200 parasites/μL of blood) was recorded at high altitude sites (Table 1 and Figure 2). No significant correlation was found between altitude and GMPD (r = −0.42, P = 0.397).
Figure 1.
Malaria prevalence at different altitude zones in the Mount Cameroon area. Error bars indicate mean ± SD.
Figure 2.
Geometric mean parasite density (GMPD) at different altitude zones in the Mount Cameroon area. Each bar represents the mean ± SD GMPD at a given altitude zone.
Entomologic indices.
Four Anopheles species were found to prevail in the region (An. gambiae, An. funestus, An. hancocki, and An. nilli). Globally, An. gambiae was found to be the most aggressive species with a high HBR (32.99 bites/person/night) in Tiko (Table 2). The highest total HBR (38.84 bites/person/night) was recorded in Mutengene, the only site in which An. funestus appeared to be a major vector with an HBR of 25.19 bites/person/night. In general, the four Anopheles species were found to be more aggressive at intermediate and low altitudes. Anopheles funestus and hancocki were more aggressive at intermediate zone and had cumulative HBRs of 35.90 bites/person/night and 5.82 bites/person/night, respectively. Anopheles gambiae was aggressive at low and intermediate altitudes (Table 2). No significant association was found between HBR and altitude (r = −0.714, P = 0.111).
Table 2.
Human-biting rates in relation to Anopheles species and locality, Mount Cameroon area
| Locality | Human biting rate (bites/person/night) | ||||
|---|---|---|---|---|---|
| An. funestus | An. gambiae | An. hancocki | An. nilli | Total | |
| Bonakanda | 0 | 0 | 0 | 0 | 0 |
| Likoko Membea | 4.38 | 1.38 | 4.92 | 0 | 10.68 |
| Meanja | 10.71 | 19.98 | 0.90 | 0 | 31.59 |
| Mutengene | 25.19 | 4.73 | 4.92 | 1.92 | 36.76 |
| Debundscha | 0 | 27.20 | 0 | 0 | 27.20 |
| Tiko | 2.54 | 32.99 | 0 | 0.67 | 36.20 |
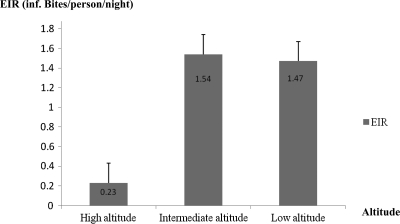
Infected An. funestus and An. hancocki were found only at high and intermediate altitudes. Infected An. gambiae was found at all altitude zones, and the highest sporozoite rate (7.36%) was recorded in Tiko (low altitude). As opposed to An. gambiae, An. nilli was found only in Mutengene and had a sporozoite rate of 7.14% (Table 3). Anopheles gambiae was the most aggressive vector and the most active vector in malaria transmission in the Mount Cameroon region (overall EIR = 3.93 infective bites/person/night). This vector had EIRs in all altitude zones, and the highest EIR (2.43 infective bites/person/night) was recorded at a low altitude (Tiko). In contrast, the three other Anopheles species had EIRs only at high and intermediate altitudes. The highest EIRs of 1.38 infective bites/person/night and 0.14 infective bites/person/night for An. funestus and An. nilli, respectively, were obtained at intermediate altitudes. The highest EIR for An. hancocki (0.25 infective bites/person/night) was recorded at a high altitude (Table 4). In all localities, An. hancocki and An. nilli were found to be minor vectors. Overall, the EIRs were higher at low and intermediate altitudes than at high altitudes. The intermediate zone appeared to have the highest EIR. The same pattern was observed with GMPD (Figures 2 and 3). However, for malaria prevalence, an unexpected lower EIR was also obtained in Debundscha (50 meters above sea level). A significant positive correlation was also found between malaria prevalence and EIR (r = 0.943, P = 0.005). As opposed to HBR, there was a significant negative correlation between EIR and altitude (r = −0.829, P = 0.04).
Table 3.
Sporozoite rates in relation to Anopheles species and locality, Mount Cameroon area
| Locality | Sporozoite rate (%) of Anopheles species | ||||
|---|---|---|---|---|---|
| An. funestus | An. gambiae | An. hancocki | An. nilli | Average | |
| Bonakanda | 0 | 0 | 0 | 0 | 0 |
| Likoko Membea | 3.96 | 3.04 | 5.19 | 0 | 3.05 |
| Meanja | 3.24 | 3.47 | 1.39 | 0 | 2.03 |
| Mutengene | 5.50 | 5.68 | 4.46 | 7.14 | 5.70 |
| Debundscha | 0 | 1.85 | 0 | 0 | 0.46 |
| Tiko | 0 | 7.36 | 0 | 0 | 1.84 |
| Average (%) | 2.12 | 3.56 | 1.84 | 1.19 | |
Table 4.
Entomologic inoculation rates in relation to Anopheles species and locality, Mount Cameroon area
| Locality | Entomologic inoculation rate (infective bites/person/night) | ||||
|---|---|---|---|---|---|
| An. funestus | An. gambiae | An. hancocki | An. nilli | Total | |
| Bonakanda | – | – | – | – | – |
| Likoko Membea | 0.17 | 0.04 | 0.25 | – | 0.46 |
| Meanja | 0.35 | 0.69 | 0.01 | 0 | 1.05 |
| Mutengene | 1.38 | 0.27 | 0.23 | 0.14 | 2.02 |
| Debundscha | 0 | 0.50 | 0 | 0 | 0.50 |
| Tiko | 0 | 2.43 | 0 | 0 | 2.43 |
| Total | 1.9 | 3.93 | 0.49 | 0.14 | |
Figure 3.
Entomologic inoculation rate (EIR) at different altitude zones in the Mount Cameroon area. Each bar represents the mean ± SD EIR at a given altitude zone.
Sampling and P. falciparum status in the Mount Cameroon region.
A total of 142 P. falciparum-positive blood samples were randomly selected for genotyping: 30 from Meanja, Mutengene, and Tiko respectively, and 19, 18, and 15 from Likoko Membea, Bonakanda, and Debundscha, respectively. Thus, we analyzed 37 samples from high altitude sites, 60 samples from intermediate altitude sites, and 45 samples from low altitude sites. Because of the discrepancy in sample size, all proportions were calculated with respect to the total number of sample tested at each altitude zone to make them comparable. Of the 142 samples genotyped, 93 (65%) were positive for MSP-1 block 2. Of the 93 positive samples, 18 (19.35%), 43 (46.24%), and 32 (34.41%) were from high, intermediate, and low altitudes sites, respectively.
Plasmodium falciparum MSP-1 block 2 allelic variant frequencies in sites of contrasting altitudes.
Among the 93 P. falciparum MSP-1 positive cases, 32 were positive for the allelic variant K-1. Six (16.21%) of 37 were from high altitude sites, 11 (18.33%) of 60 from intermediate sites, and 15 (33.33%) of 45 from low altitude sites. There was a significant difference in the frequency distribution of this allelic variant between study sites (P = 0.021). Sixty-nine of 93 samples were positive for the allelic variant MAD20. Nine (24.32%), 36 (60%), and 24 (54.33%) samples were from sites at high, intermediate, and low altitudes, respectively. There was also a significant difference in the frequency distribution of the allelic variant MAD20 between study sites (P < 0.001). Fifty samples were positive for the allelic variant RO33. Ten (27.02%), 24 (40%), and 16 (35.55%) were observed at high, intermediate, and low altitudes, respectively. The difference in the frequency distribution of the allelic variant RO33 between the study sites was not significant (P = 0.282).
Merozoite surface protein 1 block 2 type genetic diversity and altitude.
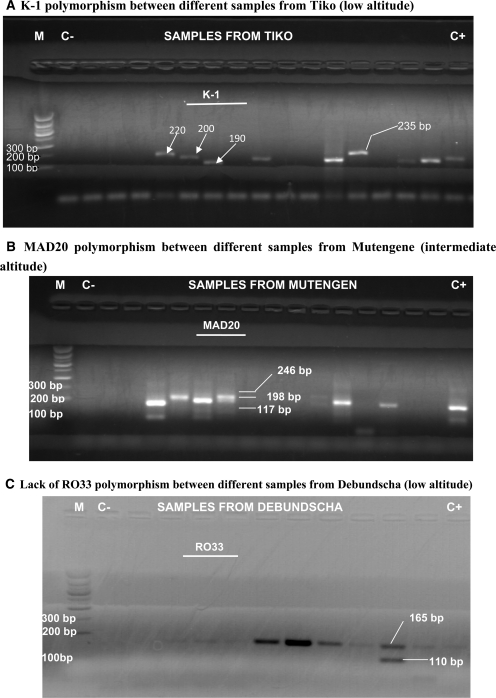
Allelic families of MSP-1 block 2 were diverse and classified according to the size of their PCR-amplified fragments. The allelic K-1 type yielded only one fragment per sample in all the localities except Debundscha (low altitude) and Mutengene and Meanja (intermediate zone) where some samples produced ≤ 2 fragments. There was a high level of polymorphism between samples that yielded one fragment (Figure 4A). Fragment sizes were within the expected range (80–459 basepairs). DNA sequencing of these K-1 PCR-products identified five DNA sequences. Database searches with the BLAST Network Service confirmed that those sequences were from the malaria parasite P. falciparum. Deduced sequence homologies are presented in Supplementary Table 1. Our sequences shared high similarity (98–100%) with K-1 sequences registered in GenBank. The fifth sequence of 250 basepairs did not match any described K-1 polymorphism. The corresponding amino acid sequence (5210B) had an atypically long region starting with SAQ and terminating with SGPSGT amino acids and consisted of 13 SGT tripeptide repeats (Figure 5). The amino acid sequence 5210B was deposited in GenBank under accession no. JF968467 as a new P. falciparum MSP-1 block 2 K-1 polymorphism identified in the Mount Cameroon malaria-endemic region.
Figure 4.
Merozoite surface protein 1 (MSP-1) block polymorphism in the Mount Cameroon area. M = molecular marker; C− = negative control; C+ = positive control. Other lanes contain the second-round polymerase chain reaction products of different samples amplified with a, K-1, b, MAD20, and c, RO33 specific primers. Bp = basepairs.
Supplementary Table 1.
NCBI BLASTanalysis of P. falciparum MSP-1 block 2 K-1 type DNA genotyping results from the Mount Cameroon area
| Sample | GenBank accession no. | K-1 allele size (bp) | BLASTN highest homology, GenBank description | E value | % Nucleotide identity | % Amino acid identity |
|---|---|---|---|---|---|---|
| 2160A | 169 | M77730.2 P. falciparum clone 834B major MSP | 9e-75 | 97 | 94 | |
| 2160A | 169 | M77730.2 P. falciparum clone 834B major MSP | 9e-75 | 97 | 94 | |
| 2210B | JF968465 | 223 | AF509633.1 P. falciparum isolate MSP-1 170 | 8e-96 | 95 | 90 |
| 190A | 178 | AF191061 P. falciparum isolate 1/M1 MSP-1 | 1e-72 | 94 | 86 | |
| 4200A | JF968466 | 205 | AB276007.1 P. falciparum msp-1 gene for MSP-1 | 1e-99 | 100 | 100 |
| 5210B | JF968467 | 250 | EU445566.1 P. falciparum type 12 MSP-1 | 4e-113 | 96 | 92 |
NCBI = National Center for Biotechnology Information; BLAST = basic local alignment search tool; MSP-1 = merozoite surface protein 1; bp = basepairs.
Figure 5.
Deduced amino acid sequences of five alleles for merozoite surface protein 2(MSP-2) block 2 K-1 type from Plasmodium falciparum isolates identified in the Mount Cameroon area. The protein alignment, which also includes the P. falciparum Thai-K1 isolate reference sequence (GenBank accession no. CAA27070), was constructed by using the MUSCLE algorithm (http://www.ebi.ac.uk/Tools/muscle/) and is shown by using the GeneDoc program (http://www.nrbsc.org/gfx/genedoc/index.html). Regions of consensus at the amino acid level are shaded and dashes represent gaps.
The allelic variant MAD20 yielded ≤ 4 four bands. The major finding here was that samples with four and three bands were mostly recorded in Debundscha, Tiko, and Mutengene (low and intermediate altitude sites) (Figure 4B). Only a limited number of samples were found to have ≤ 4 bands in Bonakanda (high altitude/low transmission). The MAD20 fragment sizes were also within the expected range (80–350 basepairs).
The allelic RO33 type was the least polymorphic. Only one sample in Debundscha resulted in amplification of ≤ 2 (Figure 4C). Most fragments were between 156 and 165 basepairs.
Multiplicity (complexity) of infection.
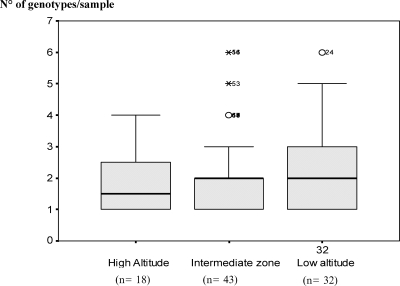
Forty-eight (54.02%) samples among which 7 (18.91%) of 37. 22 (36.66%) of 60, and 19 (42.22%) of 45 samples from high, intermediate and low altitude sites, respectively, contained multiclonal infections. There was a trend of increasing proportion of multiple-type infections with a decrease in altitude. The overall estimated mean ± SD number of clones per sample was 2.08 ± 1.34. There was also a trend of increasing mean ± SD number of clones per sample with a decrease in altitude. A total of 1.58 ± 0.8, 2.1 ± 1.24, and 2.32 ± 1.47 clones per sample were recorded in high, intermediate, and low altitude areas, respectively Up to six genotypes were found per sample at low (high malaria transmission) at intermediate altitudes; at high altitude (low malaria transmission), most samples had 2–3 genotypes (Figure 6). The Kruskal-Wallis test showed no significant differences between median numbers of clones per altitude zone (H = 0.632, P = 0.729). Similar to the Kruskal-Wallis test, the Dunn test did not detect any significant differences in multiplicity of infection between high and low altitudes (Q = 0.795, P > 0.05), high and intermediate altitudes (Q = 0.123, P > 0.05), and low and intermediate altitudes (Q = 0.981, P > 0.05).
Figure 6.
Multiplicity of infection at different altitude zones in the Mount Cameroon area. Error bars indicate mean ± SD.
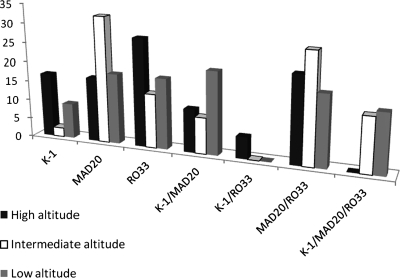
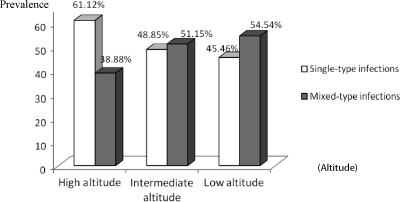
At high altitude, of the 18 positive samples obtained, single-type infections accounted for 61.12% and samples containing RO33 were most prevalent. The proportion of mixed-type infections was 38.88% and samples containing MAD 20 and RO33 prevailed. However, no samples were found with the three allelic variants (Figures 7 and 8). At intermediate altitude, of 43 samples amplified, 51.15% had mixed-type infections. Single-type infections with MAD 20 (27.77%) and mixed-type infections with MAD 20/RO33 (27.9%) were more common. No mixed-type infections with K-1/RO33 were observed at this level (Figure 7). At low altitude, of the 32 samples that were positive (amplified), 54.54% had mixed-type infections. Single and mixed-type infections were similarly represented at low altitude except that single-type infections with K-1 were less frequent. No mixed-type infections with K-1/RO33 were seen at this altitude (Figure 7).
Figure 7.
Comparison of single- and mixed-type infections at different altitude zones in the Mount Cameroon area. Percentages were calculated with respect to total positive samples at each altitude zone.
Figure 8.
Proportions of single and mixed-type infections at high attitude, intermediate altitude, and low altitude in the Mount Cameroon area. Proportions were calculated with respect to total positive samples at each altitude zone.
The Spearman correlation rank test showed no significant association between altitude and complexity of infection (r = −0.6, P = 0.208) or between EIR and multiplicity of infection (r = 0.486, P = 0.329). However, just as higher EIRs were recorded at low and intermediate zones, infections appeared to be more complex in these zones.
Discussion
This is the first report on the genetic diversity of P. falciparum field isolates from sites of contrasting altitudes or sites of contrasting malaria transmission profiles in the Mount Cameroon area in the South-West region of Cameroon. It is of great importance to understand the molecular characteristics of the parasite population because they can provide information for vaccine development.
This study demonstrated that the altitude of an area has an influence on prevalence and intensity of malaria infection. There was a decrease in the prevalence of malaria infection with an increase in altitude, and the difference at various altitudes was significant. A correlation test also showed a negative association between prevalence and altitude, although not significant. This finding is consistent with that of another study conducted in Tanzania, in which prevalence of malaria parasites decreased with an increase in altitude.23 There was also a strong negative association between altitude and EIR. This finding indicates that malaria transmission decreases with altitude and can also explain malaria prevalence rates obtained in this study. However, the unexpected high malaria prevalence observed in Meanja (intermediate altitude) could be explained by a dam that created an ecosystem appropriate for Anopheles mosquito vector development. Conversely, low malaria prevalence recorded in Debundscha (low altitude) could be attributed to rainfall in the locality. Excessive rainfall probably decreases vector activity hence low malaria transmission and prevalence. This finding is confirmed by the low EIR in the locality.
In general, it can be suggested from the EIR and sporozoite rates of different Anopheles species found in this study that An. gambiae plays a major role in malaria transmission at low altitude. At intermediate and high altitudes, An. funestus and to a lesser extent An. hancocki and An. nilli are also found, but they appear to be minor vectors. The EIR reflected the HBR; low and intermediate altitudes had higher values than high altitudes. This finding could be explained by the fact that the mosquito population decreases with altitude.26 The mosquito population is reduced at high altitude because of the steepness of the slope and absence of streams or stagnant pools of water that normally play a major role in creation of breeding sites. This zone is also characterized by low temperature and low humidity, which are not favorable for mosquito vector development. The EIR at different altitude zones (Figure 3) and the GMPD (Figure 2) also showed the same pattern; the intermediate zone had relative higher values. This finding might imply an association between malaria infection intensity in the host and that in the vector. Despite the relative higher EIR value obtained in the intermediate zone, a significant negative correlation was found between EIR and altitude. This finding corroborates results obtained in Tanzania23 because in our study, a significant positive correlation was found between EIR and prevalence.
The study also demonstrates that genetic diversity of P. falciparum may be a function of transmission (the relationship is non-linear). This result is similar to those of studies conducted in Thailand.10,14 In general, the P. falciparum MSP gene was similarly represented in all sites included in this study. Some samples failed to amplify, which might be caused by low recovery of P. falciparum DNA during DNA extraction. Furthermore, because the MSP gene seems to be polymorphic, single-nucleotide polymorphisms might be present in regions for which primers were designed and this may hinder proper primer annealing.
The three allelic variants K-1, MAD20, and RO33 were observed in all six localities. However, allelic variant K-1 was highly distributed in low altitude sites compared with intermediate and high altitude sites, but MAD20 and RO33 allelic variants were common at an intermediate altitude than at a low altitude (Table 5). This low distribution of K-1 at a high altitude could be accounted for by the low number of samples used, but using the sample size at each altitude zone in calculating the different proportions did not result in a different distribution pattern. If one considers that K-1 was rarely observed in all the localities in the Mount Cameroon region, it could be suggested that certain P. falciparum strains had single nucleotide polymorphisms in DNA regions to which primers were designed, which interfered with proper annealing for this particular allelic type. If not true, it might suggest a relative negative natural selection in the region because overall distribution of K-1 in the region was low. However, this observation does not corroborate most findings in areas of holoendemic, mesoendemic, and hyperendemic malaria, in which the allelic variant K-1 was predominant.8,15,35
Table 5.
Frequencies of merozoite surface protein 1 allelic variants with respect to locality, Mount Cameroon area
| Characteristic | Allelic type, no. (%) | |||
|---|---|---|---|---|
| K-1 | MAD20 | RO33 | ||
| High altitude | Bonakanda (18) | 3 | 8 | 6 |
| Likoko Membea (19) | 3 | 1 | 4 | |
| Total (37) | 6 (16.21) | 9 (24.32) | 10 (27.02) | |
| Intermediate altitude | Meanja (30) | 2 | 19 | 16 |
| Mutengene (30) | 9 | 17 | 8 | |
| Total (60) | 11 (18.33) | 36 (60) | 24 (40) | |
| Low altitude | Debundscha (15) | 3 | 9 | 8 |
| Tiko (30) | 12 | 15 | 8 | |
| Total (45) | 15 (33.33) | 24 (53.33) | 16 (35.55) | |
| Overall total of each allelic type (142) | 32 (22.53) | 69 (48.59) | 50 (35.21) | |
| Chi-square test | χ2 = 7.731, P = 0.021 | χ2 = 15.956, P < 0.001 | χ2 = 2.529, P = 0.282 | |
Conversely, The MAD20 allelic variant frequency was high in all sites and the same trend was observed for allelic variant combinations containing this allelic variant. This result is consistent with those of other studies. Snounou and others14 also found a high prevalence of the allelic variant MAD20 in comparison with K-1 and RO33 allelic variants. Other studies conducted in Colombia, southern Vietnam, the Brazilian Amazon, and some countries in Africa such as the Gambia, Nigeria, and Gabon reported the predominance of MAD20 and RO33 allelic variants.36–40 These findings were attributed to a positive natural selection for these two allelic variants in P. falciparum. However, these results cannot explain the result obtained in this study with certainty because the high frequency of MAD 20 we observed is with respect to the frequency of K-1, which may be caused by poor annealing as explained. Another peculiarity to our study is that RO33 allelic variant, which has been shown in many studies to be the least predominant, was the second most prevalent allelic variant type. Nevertheless, this finding corroborates findings of other studies.36–40 The high frequency of the RO33 allelic variant in the region may be associated with reduced risk of clinical malaria.41 Most patients who have this parasite allelic variant had high parasitemias (≤ 29,520 parasites/μL). However, children did not show any clinical manifestations of malaria.
Moreover, most samples with multiple polymorphisms within allelic variants (K-1 and MAD20) were obtained from low altitude sites. Even the one sample that had two polymorphisms for the allelic variant RO33 was from Debundsha (low altitude). Genetic diversity of P. falciparum populations seems to be positively associated with high parasite transmission in these geographic locations.8,12,42 In our study, sizes of MSP-1 allelic variants were within ranges obtained by other authors.43
DNA sequencing and BLAST analysis confirmed that polymorphisms obtained from PCR typing of MSP-1 allelic variant K-1 were from P. falciparum. Among the five K-1 polymorphisms obtained from P. falciparum DNA samples from the Mount Cameroon region; four had 98–100% homology with described K-1 polymorphisms. Sequences in GenBank were from P. falciparum isolates from several countries in Africa. Therefore, a vaccine based on MSP-1 allelic variant K-1 may give immune protection against malaria in those regions in Africa. However, the fifth K-1 sequence identified as 5210B-T7, which was different from any of the described K-1 sequences, could be a new polymorphism of K-1. This polymorphism could also be the basis of a malaria vaccine. However, for such an assertion to be made, further immunologic characterization of this candidate polymorphism needs to be conducted. In addition, sequencing and immunologic characterization of other allelic variants (MAD20 and R033) should also be conducted to obtain more useful information.
Detection of more than one allelic type for block 2 provided evidence for multiple P. falciparum infections of many persons on the basis of block 2 typing. Forty-eight (54%) samples were found to carry multiple allelic variants ranging from two to six distinguishable polymorphisms per infection. The overall mean ± SD number of genotypes per sample was 2.08 ± 1.34, which was similar to that observed in school children in Molyko and Buea (other localities in the Mount Cameroon region).35 Although there was no significant difference in the median numbers of genotypes/sample, there was a trend of decreasing mean number of P. falciparum genotypes or clones per person and of proportions of samples with mixed-type infections with an increase in altitude in the Mount Cameroon region. Persons living in intermediate and low altitude sites had ≤ 6 polymorphisms of P. falciparum of different allelic variants, and persons living in high altitude sites had 2–3 polymorphisms. No mixed-type infection harboring the three allelic variants was found at high altitude; this finding might reflect low transmission that prevails at that level. However, given the sample size used, a clear conclusion cannot be made. Moreover, there was a significant negative correlation between altitude and the EIR. This finding is consistent with other studies that demonstrated a decrease in malaria transmission with an increase in altitude,23,44 which in turn affects the genetic diversity of the parasite population.20,42
Genetic diversity is caused by recombination events that occur during sexual reproduction and asexual multiplication in the vector, but these recombination events are rare between allelic types. This finding can justify the high complexity of infection observed at low and intermediate altitudes where malaria transmission also appears to be high. However, the relationship between malaria transmission and multiplicity of infection is not linear. In our study, the complexity of infection was not directly associated with the level of transmission. For instance, the infection was more complex (2.32 polymorphisms/infection) at low altitude, which had an EIR of 1.47 infective bites/person/night than at intermediate altitude (2.1 polymorphisms/person), which had a relatively higher EIR of 1.54 infective bites/person/night. This finding indicates that an increase in transmission intensity does not necessarily translate into a parallel increase in the number of polymorphisms.10,45 A precise analysis of the influence of transmission intensity on the extent of parasite diversity is complicated by the marked annual variation of the EIR.46 Moreover, additional factors such as resistance to antimalaria drugs and migration (movement of older persons from one locality to another) have been shown to increase the diversity of parasite population through introduction of foreign polymorphisms.10,14,45,47 This finding could explain the relatively high complexity found in Bonakanda (1,197 meters above sea level), which in our study was characterized by an EIR close to zero. Mosquito vectors at this altitude are rare because the steepness of the slope, and the absence of streams at this altitude prevent formation of breeding sites. In addition, low temperature (< 15°C) and low relative humidity interfere with proper development of the mosquito vector.26 However with the limited sample size, a clear conclusion cannot be drawn because the relatively high complexity can be accounted for by the limited number of samples tested.
This study has demonstrated that altitude of an area has an influence on the level of endemicity and the genetic diversity of the local malaria parasite. Anopheles gambiae appeared to be the major malaria vector at low altitudes. At intermediate and high altitudes, An. funestus (which predominates at intermediate zone), An. hancocki, and An. nilli are also found, but they play minor roles in malaria transmission. Our study showed a positive association between low altitude where malaria transmission is high and MSP-1 block 2 polymorphism and complexity of infection. However, the uneven allelic polymorphisms observed in this study between different sites may be a hindrance for malaria vaccine formulations if not considered in vaccine development studies. More immunologic studies are needed to characterize the new allelic K-1 polymorphism and determine its potential to serve as a suitable vaccine candidate.
ACKNOWLEDGMENTS
We thank the persons who willingly consented to participate in this study, and Dr. G. Snounou for providing the research team with P. falciparum MSP-1 block 2 positive controls.
Footnotes
Financial support: This study was supported by the Research Foundation of Tropical Diseases and the Environment.
Authors' addresses: Samuel Wanji, Arnaud J. Kengne-Ouafo, Nicholas Tendongfor, and David D. Sofeu-Feugaing, Research Foundation for Tropical Diseases and the Environment, Buea, South West Region, Cameroon; and Department of Biochemistry and Microbiology, Faculty of Science, University of Buea, Buea, South West Region, Cameroon, E-mails: swanji@yahoo.fr, arnaudkengne@yahoo.com, ntendongfor@yahoo.com, and sofeu.feugaing@ubuea.cm. Ebanga E. Joan Eyong and Judith L. Ndamukong-Nyanga, Research Foundation for Tropical Diseases and the Environment, Buea, South West Region, Cameroon; and Department of Plant and Animal Sciences, Faculty of Science, University of Buea, Buea, South West Region, Cameroon, E-mails: jeebanga@yahoo.com and ndamju@yahoo.com. Helen K. Kimbi, Department of Plant and Animal Sciences, Faculty of Science, University of Buea, Buea, South West Region, Cameroon, E-mail: hkimbi@yahoo.co.uk. Hugues C. Nana-Djeunga, General Biology Laboratory, Department of Animal Biology and Physiology, Faculty of Science, University of Yaounde 1, Yaounde, Cameroon, E-mail: nanaclotaire@yahoo.com. Catherine Bourguinat, Institute of Parasitology, McGill University, Sainte Anne-de-Bellevue, Quebec, Canada, E-mail: catherine.bourguinat@mail.mcgill.ca. Claude L. Charvet, Institut National de la Recherche Agronomique, UR1282 Infectiologie Animale et Santé Publique, Nouzilly, France, E-mail: claude.charvet@tours.inra.fr.
References
- 1.World Health Organization, United Nations Children's Fund . World Malaria Report. 2005. http://www.who.int.org Available at. Accessed April 12, 2006. [Google Scholar]
- 2.Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. 2004;4:169–180. doi: 10.1038/nri1311. [DOI] [PubMed] [Google Scholar]
- 3.Trigg K. In: The current global malaria situation: Malaria: Parasite Biology, Pathogenesis and Protection. Sherman IW, editor. Washington, DC: American Society for Microbiology Press; 1998. p. 575. [Google Scholar]
- 4.Egan A. Focus 5 Brief 4 of 11. Washington, DC: International Food Policy Research Institute; 2001. (Malaria in Health and Nutrition: Emerging Re-emerging Issues in Developing Countries). [Google Scholar]
- 5.Titanji PV, Tamu VD, Akenji TK, Joutshop AS. Immunoglobulin G and subclass responses to Plasmodium falciparum antigens: a study in highly exposed Cameroonians. Clin Chem Lab Med. 2002;40:937–940. doi: 10.1515/CCLM.2002.164. [DOI] [PubMed] [Google Scholar]
- 6.Achidi EA, Ajua A, Kimbi KH, Sinju CM. In vivo efficacy study of quinine sulphate in the treatment of uncomplicated P. falciparum malaria in patients From South Western Cameroon. East Afr Med J. 2005;82:181–185. doi: 10.4314/eamj.v82i4.9278. [DOI] [PubMed] [Google Scholar]
- 7.Kimbi HK, Nkuo-Akenji TK, Patchhong AF, Ndamukong KN, Nkwescheu A. The comparative efficacy of malartin, with and without amodiaquine, in the treatment of Plasmodium falciparium malaria in the Buea district of Cameroon. Ann Trop Med Parasitol. 2006;101:1–8. doi: 10.1179/136485907X156942. [DOI] [PubMed] [Google Scholar]
- 8.Konate L, Zwetyenga J, Rogier C, Bischoff E, Fontenille D, Tall A, Spiegel A, Trape J F, Mercereau-Piujalon O. The epidemiology of multiple Plasmodium falciparum infections. 5. Variations in Plasmodium falciparum MSP-1 block 2 and MSP-1 allele prevalence and of infection complexity in two neighbouring Senegealese villages with different transmission conditions. Trans R Soc Trop Med Hyg. 1999;93:21–28. doi: 10.1016/s0035-9203(99)90323-1. [DOI] [PubMed] [Google Scholar]
- 9.Kiwanuka GN. Genetic diversity in Plasmodium falciparum merozoite surface protein 1 and 2 coding genes and its implications in malaria epidemiology: a review of published studies from 1997–2007. J Vector Borne Dis. 2009;46:1–12. [PubMed] [Google Scholar]
- 10.Paul RE, Hackford I, Brockman A, Muller-Graf C, Price R, Luxemburger C, White NJ, Nosten E, Day PK. Transmission intensity and Plasmodium falciparum diversity on the northwestern border of Thailand. Am J Trop Med Hyg. 1998;58:195–203. doi: 10.4269/ajtmh.1998.58.195. [DOI] [PubMed] [Google Scholar]
- 11.Babiker HA, Ranford-Cartwright LC, Walliker D. Genetic structure and dynamics of Plasmodium falciparum infections in the Kilombero Region of Tanzania. Trans R Soc Trop Med Hyg. 1999;93:11–14. doi: 10.1016/s0035-9203(99)90321-8. [DOI] [PubMed] [Google Scholar]
- 12.Bendixen M, Msangeni HA, Pedersen BV. Diversity of Plasmodium falciparum population and complexity of infections in relation to transmission and host age: a study from the Usambara Mountains, Tanzania. Trans R Soc Trop Med Hyg. 2001;95:143–148. doi: 10.1016/s0035-9203(01)90140-3. [DOI] [PubMed] [Google Scholar]
- 13.Kang J, Moon S, Kim J, Cho S, Lin K, Sohn W, Kim T, Na B. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum field isolates from Myanmar. Malar J. 2010;9:131–139. doi: 10.1186/1475-2875-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp 1 and msp 2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- 15.Peyerl-Hoffmann G, Jelinek T, Kilian A, Kabagambe G, Metzger WG, Sonnenburg FV. Genetic diversity of Plasmodium falciparum and its relationship to parasite density in an area with different malaria endemicities in west Uganda. Trop Med Int Health. 2001;6:607–613. doi: 10.1046/j.1365-3156.2001.00761.x. [DOI] [PubMed] [Google Scholar]
- 16.Tanabe K, Mackay M, Goman M, Scaife JG. Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J Mol Biol. 1987;195:273–287. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 17.Ferreira MU, Liu Q, Kaneko O, Kimura M, Tanabe K, Kimura EA, Katzin M, Isomura S, Kawamoto F. Allelic diversity at the merozoite surface protein-1 locus of Plasmodium falciparum in clinical isolates from the South-Western Brazilian Amazon. Am Soc Trop Med Hyg. 1998;59:474–480. doi: 10.4269/ajtmh.1998.59.474. [DOI] [PubMed] [Google Scholar]
- 18.Da-Silveira AL, Dorta LM, Kimura SA, Katzin MA, Kawamoto F, Kazuyuki T, Ferreira UM. Allelic diversity and antibody recognition of Plasmodium falciparum merozoite surface protein-1 during hypoendemic malaria transmission in the Brazilian Amazon region. Infect Immun. 1999;67:5906–5916. doi: 10.1128/iai.67.11.5906-5916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller LH, Roberts T, Shahabuddin M, McCutchan TF. Analysis of sequence diversity in the Plasmodium falciparum merozoite surface protein-1 (MSP-1) Mol Biochem Parasitol. 1993;59:1–14. doi: 10.1016/0166-6851(93)90002-f. [DOI] [PubMed] [Google Scholar]
- 20.Kerr PJ, Randford-Cartwright LC, Walliker D. Proof of intragenic recombination in Plasmodium falciparum. Mol Biochem Parasitol. 1994;66:241–248. doi: 10.1016/0166-6851(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 21.Farnert A, Rooth I, Svensson AK, Snounou G, Bjorkmane A. Complexity of Plasmodium falciparum infections is consistent over time and protects against clinical disease in Tanzanian children. J Infect Dis. 1999;179:989–995. doi: 10.1086/314652. [DOI] [PubMed] [Google Scholar]
- 22.Takala S, Branch O, Escalante AA, Kariuki S, Wootton J, Lal AA. Evidence for intragenic recombination in Plasmodium falciparum: identification of a novel allele family in block 2 of merozoite surface protein-1: Asembo Bay Area Cohort Project XIV. Mol Biochem Parasitol. 2002;125:163–171. doi: 10.1016/s0166-6851(02)00237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drakeley CJ, Carneiro I, Reyburn H, Malima R, Lusingu JP, Cox J, Theander TG, Nkya WM, Lemnge MM, Riley EM. Altitude-dependent and independent variations in Plasmodium falciparum prevalence in northeastern Tanzania. J Infect Dis. 2005;191:1589–1598. doi: 10.1086/429669. [DOI] [PubMed] [Google Scholar]
- 24.Akenji TN, Ntonifor NN, Ching JK, Kimbi HK, Ndamukong KN, Anong DN, Boyo MG, Titanji VP. Evaluating a malaria intervention Strategy using knowledge practices and coverage surveys in rural Bolifamba, SouthWest Cameroon. Trans R Soc Trop Med Hyg. 2005;99:325–332. doi: 10.1016/j.trstmh.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 25.Kimbi HK, Nformi D, Ndamukong KJ. Prevalence of asymptomatic malaria among school children in an urban and rural area in the Mount Cameroon region. Cent Afr Med J. 2005;51:5–10. [PubMed] [Google Scholar]
- 26.Wanji S, Tanke T, Tanga SN, Ajonina C, Tendongfor N, Fontenille D. Anopheles species of the Mount Cameroon regions: biting habits, feeding behaviour and entomological inoculation rates. Trop Med Int Health. 2003;8:643–649. doi: 10.1046/j.1365-3156.2003.01070.x. [DOI] [PubMed] [Google Scholar]
- 27.Cheesbrough M. District Laboratory Practice in Tropical Countries. Part 1. London: Cambridge University Press; 2000. p. 454. [Google Scholar]
- 28.World Health Organization . Bench Aids for the Diagnosis of Malaria. Geneva: World Health Organization; 2000. [Google Scholar]
- 29.Gilles MT, De Meillon B. The Anophelines of Africa South of the Sahara (Ethiopian Zoogeogaphical Region) Second edition. Johannesburg, South Africa: South African Institute for Medical Research; 1968. p. 54. [Google Scholar]
- 30.Gilles MT, Coetzee M. A Supplement to the Anophelinae of Africa South of the Sahara. Johannesburg, South Africa: South African Institute for Medical Research; 1987. p. 55. [Google Scholar]
- 31.Harvey JP, Le Goff G, Geoffroy G, Herve JP, Manga L, Brunches J. Les Anopheles de la Région Afro-Tropical. Paris: Français de Recherché Scientifique pour le Développement en Coopération et Institut de Recherche pour le Développement; 1998. [Google Scholar]
- 32.Nkondjio AC, Awono AP, Teto JC, Meuni Y, Zebaze KS, Nyambam R. High malaria transmission intensity in a village close to Yaounde capital city of Cameroon. J Med Entomol. 2002;39:350–355. doi: 10.1603/0022-2585-39.2.350. [DOI] [PubMed] [Google Scholar]
- 33.Hamad AA, Nugud AE, Arnot DE, Giha HA, Abdel-Muhsin AA, Satti GM, Theander TG, Creasey AM, Babiker HA, Elnaiem DA. A marked seasonality of malaria transmission in two rural sites in eastern Sudan. Acta Trop. 2002;83:71–82. doi: 10.1016/s0001-706x(02)00059-1. [DOI] [PubMed] [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimbi HK, Tetteh AK, Polley SD, Conway DJ. Cross sectional study of specific antibodies to a polymorphic Plasmodium falciparum antigen and of parasite antigen genotypes in school children on the slope of mount Cameroon. Trans R Soc Trop Med Hyg. 2004;98:284–289. doi: 10.1016/S0035-9203(03)00068-3. [DOI] [PubMed] [Google Scholar]
- 36.Snewin VA, Herrera M, Sanchez G, Scherf A, Langsley G, Herrera S. Polymorphism of the alleles of the merozoite surface antigens MSA1 and MSA2 in Plasmodium falciparum wild isolates from Colombia. Mol Biomed Parasitol. 1991;49:265–276. doi: 10.1016/0166-6851(91)90070-m. [DOI] [PubMed] [Google Scholar]
- 37.Ferreira MU, Kaneko O, Masatsugu K, Qing L, Kawamoto F, Tanabe K. Allelic diversity at the merozoite surface protein-1 (MSP-1) locus in natural Plasmodium falciparum populations: a brief overview. Mem Inst Oswaldo Cruz. 1998;93:631–638. doi: 10.1590/s0074-02761998000500013. [DOI] [PubMed] [Google Scholar]
- 38.Gomez D, Chaparro J, Rubiano C, Orfa RM, Wasserman M. Genetic diversity of Plasmodium falciparum field samples from isolated Colombian village. Am Soc Trop Med Hyg. 2002;67:611–616. doi: 10.4269/ajtmh.2002.67.611. [DOI] [PubMed] [Google Scholar]
- 39.Ferreira MU, Ribiero WL, Tonon AP, Kawamoto F, Rich SM. Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Genet. 2003;304:65–75. doi: 10.1016/s0378-1119(02)01180-0. [DOI] [PubMed] [Google Scholar]
- 40.Terrientes ZI, Vergara J, Kramer K, Herrera S, Changa SP. Restricted genetic diversity of Plasmodium falciparum major merozoite surface protein 1 in isolates from Colombia. Am J Trop Med Hyg 73 (Suppl): 2005:55–61. doi: 10.4269/ajtmh.2005.73.55. [DOI] [PubMed] [Google Scholar]
- 41.Al-Yaman F, Genton B, Reeder JC, Anders RF, Smith T, Alpers MP. Reduced risk of clinical malaria in children infected with multiple clones of Plasmodium falciparum in a highly endemic area: a prospective community study. Trans R Soc Trop Med Hyg. 1997;60:1056–1060. doi: 10.1016/s0035-9203(97)90046-8. [DOI] [PubMed] [Google Scholar]
- 42.Babiker HA, Lines J, Hill WG, Walliker D. Population structure of Plasmodium falciparum in villages with different malaria endemicity in east Africa. Am J Trop Med Hyg. 1997;56:141–147. doi: 10.4269/ajtmh.1997.56.141. [DOI] [PubMed] [Google Scholar]
- 43.Najia KG, Martensson J, Ursing A, Jafri S, Bereczky S, Hussain R, Beg MA. Genetic diversity among Plasmodium falciparum field isolates in Pakistan measured with PCR genotyping of the merozoite surface protein 1 and 2. Malar J. 2010;9:1. doi: 10.1186/1475-2875-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Githeko AK, Ayisi JM, Odada PK, Atieli FK, Ndenga BA, Githure JI, Yan G. Topography and malaria transmission heterogeneity in western Kenya highlands: prospects for focal vector control. Malar J. 2006;5:107. doi: 10.1186/1475-2875-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zwetyenga J, Rogier C, Tall A, Fontenille D, Snounou G, Trape JF, Mercereau-Puijalon O. No influence of age on infection complexity and allelic distribution in Plasmodium falciparum infections in Ndiop, a Senegalese village with seasonal, mesoendemic malaria. Trans R Soc Trop Med Hyg. 1998;59:726–735. doi: 10.4269/ajtmh.1998.59.726. [DOI] [PubMed] [Google Scholar]
- 46.Fontenille D, Lochouarn L, Diatta M, Sokhna C, Dia I, Diagne N, Lemasson JJ, Ba K, Rogier C, Tall A, Trape JF. A four year entomological study of the transmission of seasonal malaria in Senegal and bionomics of Anopheles gambiae and A. arabiensis. Trans R Soc Trop Med Hyg. 1997;91:647–652. doi: 10.1016/s0035-9203(97)90506-x. [DOI] [PubMed] [Google Scholar]
- 47.Zhong D, Afrane Y, Githeko A, Yang Z, Cui L, Menge DM, Temu EA, Yan G. Plasmodium falciparum genetic diversity in western Kenya highlands. Am J Trop Med Hyg. 2007;77:1043–1050. [PubMed] [Google Scholar]