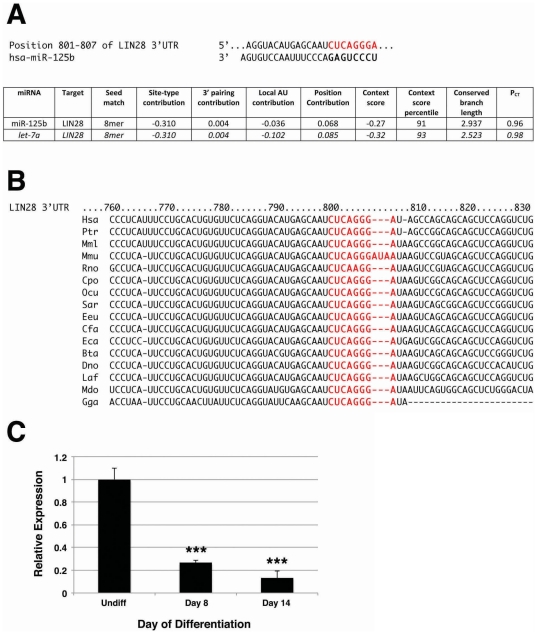
Figure 4. Lin28 is a predicted target of miR-125b in differentiating hESCs.
A) In silico analysis of potential miR-125b targets using Target Scan Human identified Lin28 as a potential target. Seed match refers to the match between a sequence in the 3′ untranslated region (UTR) of the putative target gene and positions 2–8 of the mature miRNA; site-type contribution reflects the average contribution of each site identified; 3′ pairing contribution describes the impact of miRNA-target complementarity outside the seed region; local AU contribution is a measure of the target transcript AU content 30 nucleotides upstream and downstream of the predicted site; position contribution reflects the distance to the nearest end of the target's UTR. For each of these criteria, a more negative score indicates a more favorable site. The context score is the sum of the contributions of site-type, 3′ pairing, local AU, and position criteria. Conserved branch length is the sum of phylogenetic branch lengths between species that contain the site. The probability of conserved targeting (PCT) reflects the Bayesian estimate of the probability that a site is evolutionarily conserved through selective advantage, rather than by chance. These criteria applied to miR-125b/Lin28 were similar to those for the established let-7a/Lin28 interaction. B) Conservation among mammals of the miR-125b binding site in the 3′ UTR of Lin28. Bta, cow; Cfa, dog; Cpo, guinea pig; Dno, armadillo; Eca, horse; Eeu, hedgehog; Gga, chicken; Hsa, human; Laf, elephant; Mdo, opossum; Mml, rhesus; Mmu, mouse; Ocu, rabbit; Ptr, chimpanzee; Rno, rat; Sar, shrew. C) Undifferentiated αMHC-GFP reporter hESCs (Undiff) and αMHC-GFP+ CMs sorted from reporter hESCs grown in differentiation medium for 8 or 14 days were evaluated for Lin28 expression by qPCR. Endogenous expression of Lin28 mRNA declined with CM differentiation. Data shown are mean±s.e.m. (N = 3). ***, p<0.001.