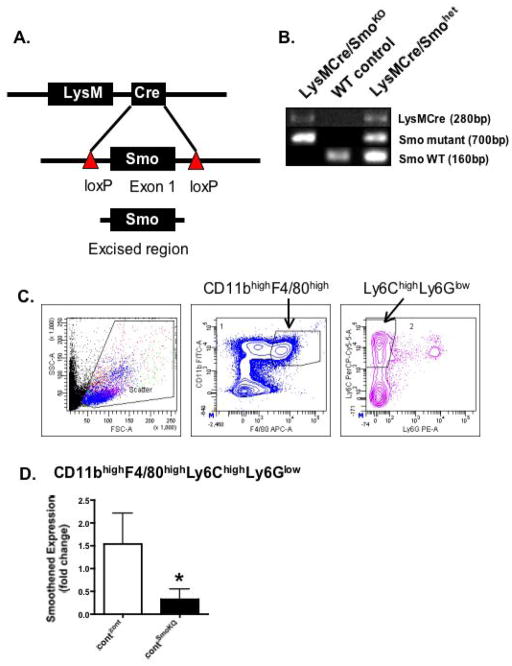
Fig. 4. Generation and verification of mice expressing a deletion of Smoothened in the myeloid cell lineage (LysMCre/SmoKO mice).
(A) A mouse model expressing a myeloid cell-specific deletion of Smoothened (LysMCre/SmoKO) was generated using transgenic mice bearing loxP sites flanking exon 1 of the Smo gene (Smo loxP) and mice expressing a Cre recombinase transgene from the Lysozyme M locus (LysMCre). (B) Genotyping was based on polymerase chain reaction primers specific for Cre, mutant Smo and wild type Smo (Smo WT). Shown are the fragments amplified from LysMCre/SmoKO, WT control and LysMCre/Smohet (heterozygous for WT and mutant Smo) mice. (C) Representative flow cytometric contour plots of the gating scheme for CD11bhighF4/80highLy6ChighLy6Glow macrophages in peripheral blood collected from control recipients transplanted with control donor (contcont) or LysMCre/SmoKO donor (contSmoKO) bone marrow cells. (D) RNA was extracted from FACS sorted macrophages and analyzed for expression of smoothened. Shown is the fold change relative to the contcont group.