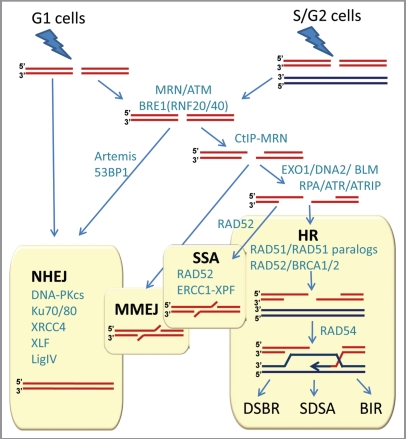
Figure 2.
Double-strand break repair in mammalian cells. NHEJ, HR, MMEJ and SSA pathways compete for the repair of DSBs. Proteins involved in various stages of DSB repair are shown in teal. References for proteins involved at different stages can be found in references 7 and 21–23. NHEJ is involved throughout the cell cycle, and HR is involved after replication, when the sister chromatid is available. A simple DSB occurring within euchromatin will be most likely repaired by NHEJ, while more complex DSBs or those in heterochromatin will initiate the full DNA damage response involving MRE11/RAD50/NBS1 (MRN) complex and ATM protein. MRN at the DSB recruits ATM, which phosphorylates BRE1A/B complex leading to relaxation of chromatin, facilitating repair by either NHEJ or HR. MRN recruits CtIP to initiate resection. Extensive resection is performed upon recruitment of EXO1 or DNA2 and the resulting single-stranded DNA (ssDNA) 3′-overhang is rapidly coated by RPA. The ssDNA-RPA recruits ATRIP leading to activation of ATR. The RPA then is displaced by Rad51 to form a nucleoprotein filament and initiate synapsis, a search for homologous template and DNA strand invasion, leading to formation of a D-loop intermediate. Synapsis is central to all HR reactions, and subsequent resolution of the D-loop is accomplished by at least three different pathways: double-strand break repair (DSBR), synthesis-dependent strand annealing (SDSA), and break-induced replication (BIR). An alternative pathway, microhomology-mediated end joining (MMEJ), which is active throughout the cell cycle, anneals short homologous sequences revealed by limited resection. Single-strand annealing (SSA) mediates repair when perfect repeats are revealed during end resection.