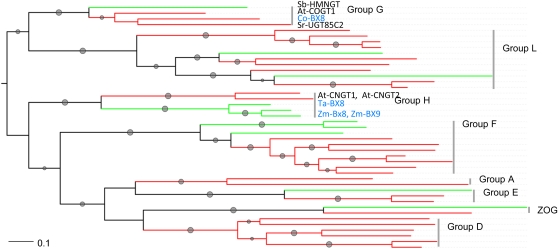
Figure 4.
Phylogenetic Tree of Plant Family 1 UGTs.
Representative enzymes with biochemically defined functions were used in the analysis. One thousand bootstrap samples were generated; branches with bootstrap values higher than 50% are marked with gray dots, and the size is proportional to the value. The trees are colored by organism: monocots in green and dicots in red. The enzymes of benzoxazinoid metabolism are written in blue letters. The affiliation to UGT groups is indicated. UGTs and accession numbers from top to bottom: Group G: HMNGT_SORBI S. bicolor, COGT2_ARATH Arabidopsis, Co-BX8 C. orientalis, Sr-UGT85C2 (UGT85C2) Stevia rebaudiana. Group L: LGT_CITUN Citrus unshiu, UGT84A10 Brassica napus, AAF98390 B. napus. IABG_MAIZE Z. mays, UGT74B1 Arabidopsis, UGT74G1 St. rebaudiana. BAA36421 Perilla frutescens, BAD06874 Iris hollandia, UGT75B2 Arabidopsis, UGT75B1 Arabidopsis, NP_188793 Arabidopsis. Group H: At-CNGT1 (CNGT1_ARATH) Arabidopsis, At-CNGT2 (CNGT2_ARATH) Arabidopsis, Ta-BX8 Triticum aestivum, Zm-BX8, Zm-BX9 Z. mays. Group F: BZ1 Z. mays, UFOG_HORVU H. vulgare, BAD83701 I. hollandica, Co-F3GT C. orientalis, Lg-F3GT L. galeobdolon, UFOG_Gentr Gentiana triflora, NP_197207 Arabidopsis, NP_564357 Arabidopsis, AAU09442 Fragaria × ananassa, UFOG_VITVI Vitis vinifera. Group A: BAD95882 Ipomoea tricolor, Bp-UGAT Bellis perennis. Group E: NM0011539 Z. mays, NP_201470 Arabidopsis, NP_566938 Arabidopsis. Family 1 UGTs of zeatin metabolism: NP_001105017 Z. mays, ZOG_PHALU Phaseolus lunatus. Group D: UGT73K1 Medicago truncatula, BAD89042 Sa-GT4 Solanum aculeatissimum, BAD29722 Q6F4D5_CATRO C. roseus, CAB56231 Dorotheanthus bellidiformis, NP_181213 Arabidopsis, NP_181218 Arabidopsis.