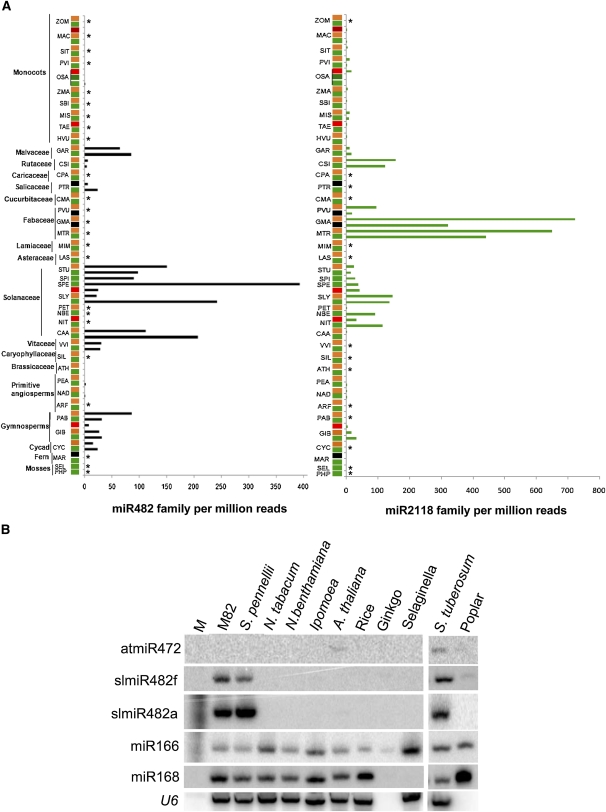
Figure 2.
Abundance of miR2118/482 Superfamily Members in Different Plants.
(A) Small RNA data sets were accessed through GEO, and miRNA abundance was analyzed through miRProf (Moxon et al., 2008) and expressed as counts per million reads. miR482 and 2118 sequences are defined in the text. Samples were from leaf/shoot tissue (green), floral tissue (orange), panicle/tassel (red), and other (black). Asterisk indicates those plant species where miRNA family members were not cloned. PHP, Physcomitrella patens; SEL, Selaginella; MAR, Marsilea; CYC, Cycas; GIB, Gingko biloba; PAB, Picea abies; ARF, Aristolochia fimbriata; NAD, Nuphar advena; PEA, Persea americana; ATH, Arabidopsis; SIL, Silene latifolia; VVI, V. vinifera; CAA, Capsicum annum; NIT, Nicotiana tabacum; NBE, N. benthamiana; PET, Petunia hybrida; SLY, S. lycopersicum; SPE, S. pennellii; SPI, S. pimpinellifolium; STU, S. tuberosum; LAS, Lactuca sativus; MIM, Mimulus; MTR, Medicago truncatula; GMA, G. max; PVU, Phaseolus vulgaris; CMA , Cucurbita maxima; PTR, Populus trichocarpa; CPA, Carica papaya; CSI, Citrus sinensis; GAR, Gossypium arboreum; HVU, Hordeum vulgare; TAE, Triticum aestivum; MIS, Miscanthes; SBI, Sorghum bicolor; ZMA, Zea mays; OSA, O. sativa; PVI, Panicum virgatum; SIT, Setaria italica; MAC, Musa acuminata; ZOM, Zostera marina.
(B) RNA gel blot analysis of tomato miR482 isoforms and cross-hybridizing homologs in different species. Fifteen micrograms of total RNA from young seedlings was used for analysis of each sample. Total RNA was electrophoresed in a 15% polyacrylamide gel, transferred to membrane, and probed with corresponding DNA oligonucleotides (see Supplemental Table 1 online) labeled with [γ-32P]ATP. U6 serves as loading control. M, decade (Ambion) size marker.