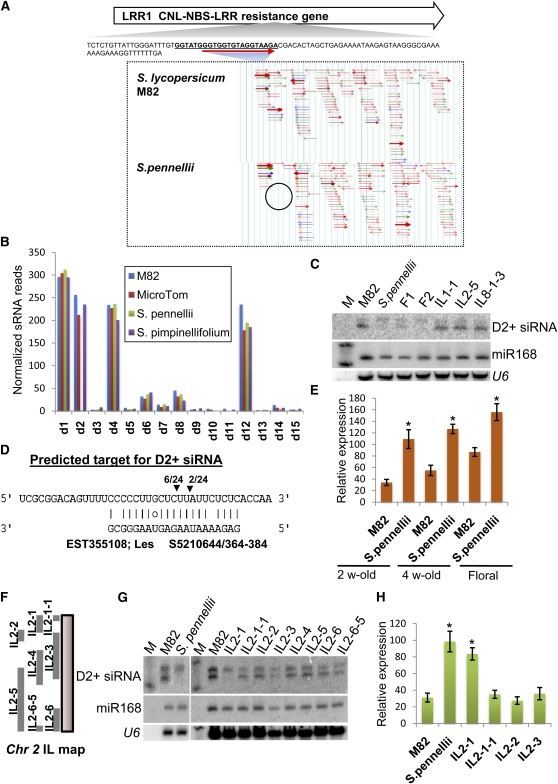
Figure 9.
Natural Variation in Secondary siRNAs Derived from LRR1.
(A) Genome browser view of secondary siRNAs aligned to LRR1 mRNA. The miR482 target site at the 5′ end of the gene is indicated. The circled region shows the absence of D2+siRNA in S. pennellii.
(B) Bar chart showing accumulation of phased secondary siRNAs in two cultivars of tomato (M82 and MicroTom) and two wild species (S. pennellii and S. pimpinellifolium). D1 to D15 indicate the different positions in the phased register that is established by the miR482 targeting event.
(C) RNA gel blot analysis of D2+ siRNA in tomato, S. pennellii, their F1 and F2 hybrids, and in introgression lines (ILs). The D2+ probe detected a 24- and 21-nucleotide species (top and bottom signal) of which only the 21-nucleotide species was absent in S. pennellii and the F1 and F2 samples. M, decade size marker.
(D) Predicted target for the D2+ siRNA derived from LRR1.
(E) Quantitative PCR analysis of abundance of D2+ target gene between M82 and S. pennellii. Error bars indicate sd (n = 6), and asterisks indicate a significant difference from corresponding control samples in S. pennellii (t test, P value < 0.05).
(F) Genetic map of IL2 series introgression lines (Eshed and Zamir, 1995). The S. pennellii genomic regions in each IL are marked.
(G) RNA gel blot analysis of D2+ siRNA in tomato, S. pennellii, and the IL2 series introgression lines.
(H) Accumulation of the D2+ target mRNA in tomato, S. pennellii, and the IL2 series introgression lines. Error bars indicate sd (n = 3), and asterisks indicate a significant difference from corresponding control samples in S. pennellii and IL2-1 (t test, P value < 0.05).