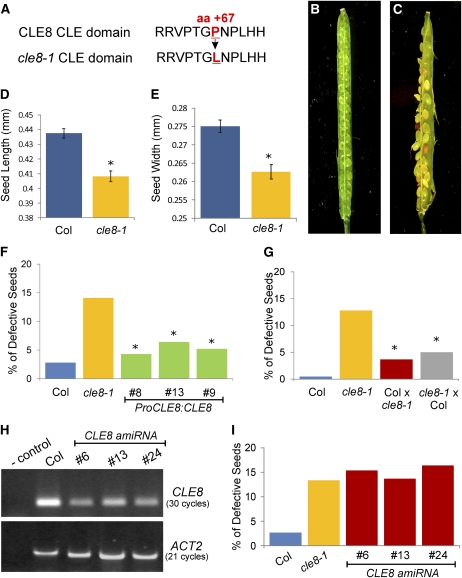
Figure 2.
cle8-1 and CLE8 amiRNA Seed Phenotypes.
(A) Wild-type CLE8 CLE domain sequence (Top) and cle8-1 CLE domain sequence (Bottom). The mutated amino acid and its position in the protein sequence are indicated in red.
(B) and (C) A wild-type (B) and a cle8-1 (C) open silique containing mature seeds. This cle8-1 silique contains more than the average number of abnormal seeds but illustrates the range of seed defects observed.
(D) and (E) Length (D) and width (E) of wild-type Col and cle8-1 seeds (n = 100).
(F) cle8-1 complementation experiment (n = 118 to 186). All transgenic lines are in the cle8-1 homozygous background. Graph values are means ± se, * indicates the difference is statistically significant (Z test, P < 0.05) when compared with the cle8-1 value but not when compared with the wild-type value.
(G) Reciprocal crosses between wild-type Col and cle8-1 plants (n = 179 to 226). * indicates the values are statistically different from wild-type and cle8-1 values (Z test, P < 0.05). The difference in the percentage of defective seeds between the two reciprocal crosses is not significant.
(H) RT-PCR analysis performed on RNA extracts from siliques of Col and three independent transgenic CLE8 amiRNA lines, using two technical replicates.
(I) Quantification of defective seeds in Col, cle8-1, and the CLE8 amiRNA transgenic lines (n = 122 to 275). The difference between wild-type Col and the other genotypes is statistically significant (Z test, P < 0.01), whereas the CLE8 amiRNA transgenic line values are not significantly different from the cle8-1 values.