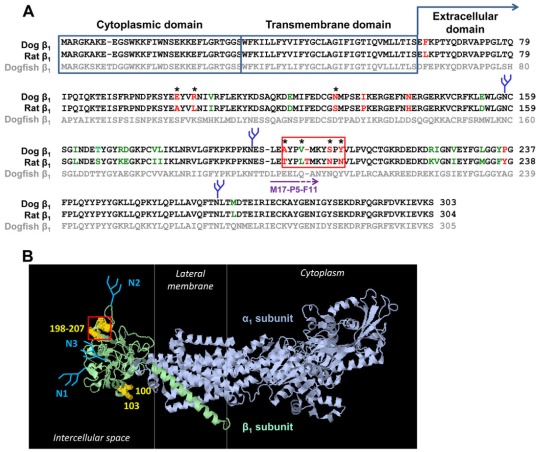
Fig. 2.
Prediction of putative β1–β1 interacting amino acid residues. (A) A computational alignment of amino acid sequences of the Na+/K+-ATPase dog β1 subunit (NP_001003283.1), rat β1 subunit (NP_037245.2) and chain B, crystal structure of the sodium-potassium pump (CAQ53919.1) was performed by using the Cobalt Multiple Alignment Tool. The amino acid residues that are identical in dog and rat subunits are shown in black font, similar amino acid residues are shown in green font, and different amino acid residues are shown in red font. The most variable region between the dog and rat sequences is highlighted by the red box. (B) The amino acid residues different in dog and rat sequences were mapped on a high-resolution structure of the Na+/K+-ATPase (2ZXE) (Shinoda et al., 2009) by using Jmol, version 1.45, to identify the species-specific residues exposed at the surface of the extracellular domain. These residues are indicated by asterisks in A and by yellow halos in B. N-glycans are shown in blue. The epitope of the M17-P5-F11 antibody is shown by a purple arrow in A.