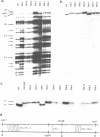
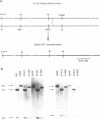
Abstract
The lin-29 gene product of C.elegans activates a temporal developmental switch for hypodermal cells. Loss-of-function lin-29 mutations result in worms that fail to execute a stage-specific pattern of hypodermal differentiation that includes exist from the cell cycle, repression of larval cuticle genes, activation of adult cuticle genes, and the cessation of molting. Combined genetic and physical mapping of restriction fragment length polymorphisms (RFLPs) was used to identify the lin-29 locus. A probe from the insertion site of a Tc1 (maP1), closely linked and to the left of lin-29 on the genetic map, was used to identify a large set of overlapping cosmid, lambda and yeast artificial chromosome (YAC) clones assembled as part of the C.elegans physical mapping project. Radiolabeled DNA from one YAC clone identified two distinct allele-specific alterations that cosegregated with the lin-29 mutant phenotype in lin-29 intragenic recombinants. lin-29 sequences were severely under-represented in all cosmid and lambda libraries tested, but were readily cloned in a YAC vector, suggesting that the lin-29 region contains sequences incompatible with standard prokaryotic cloning techniques.
Full text
PDF

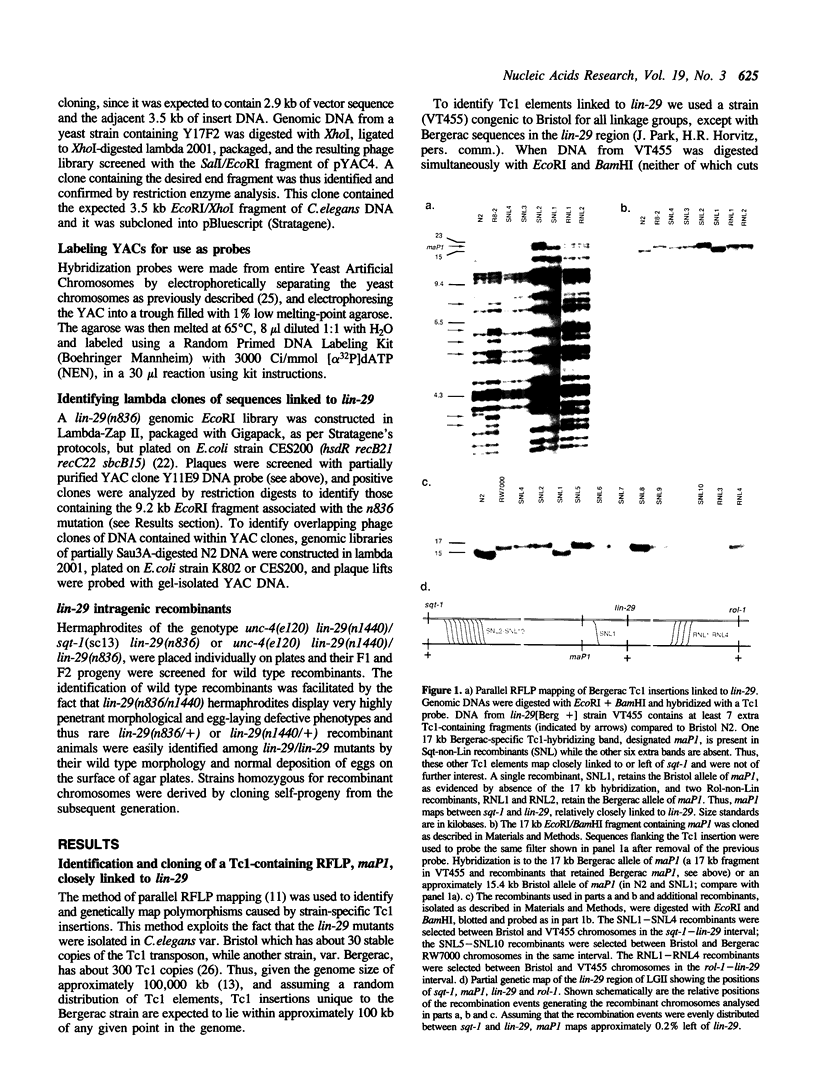
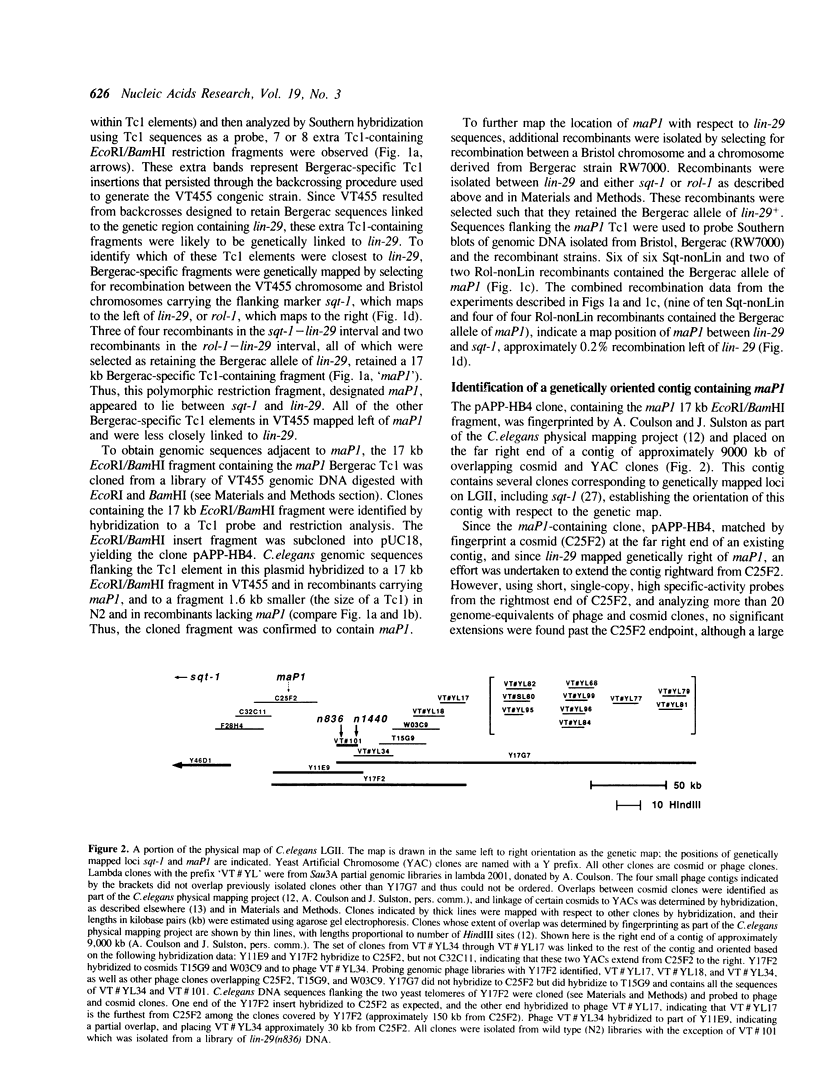
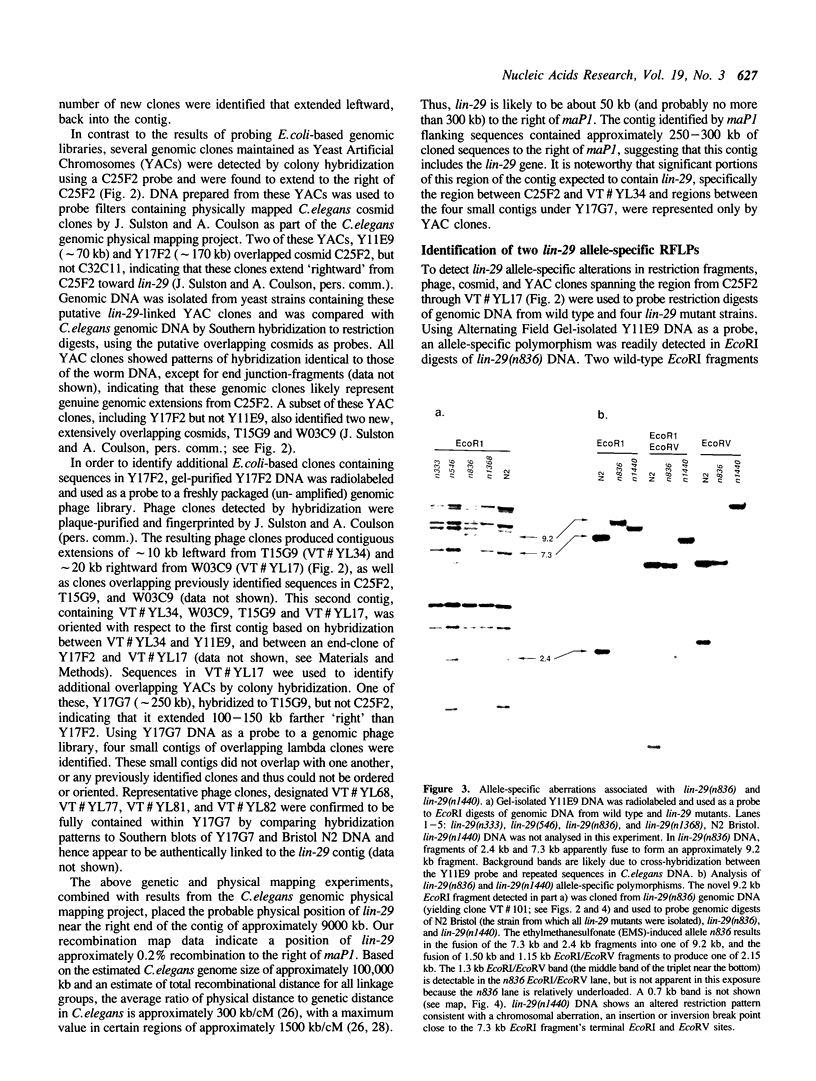
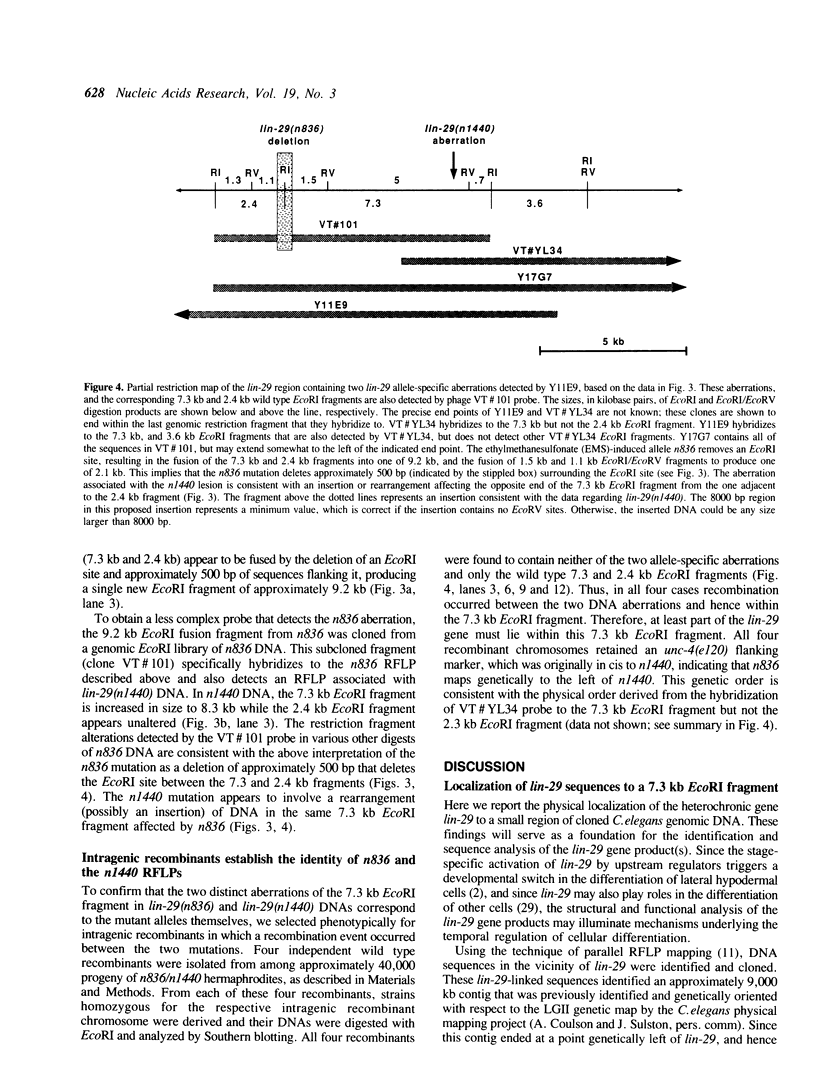
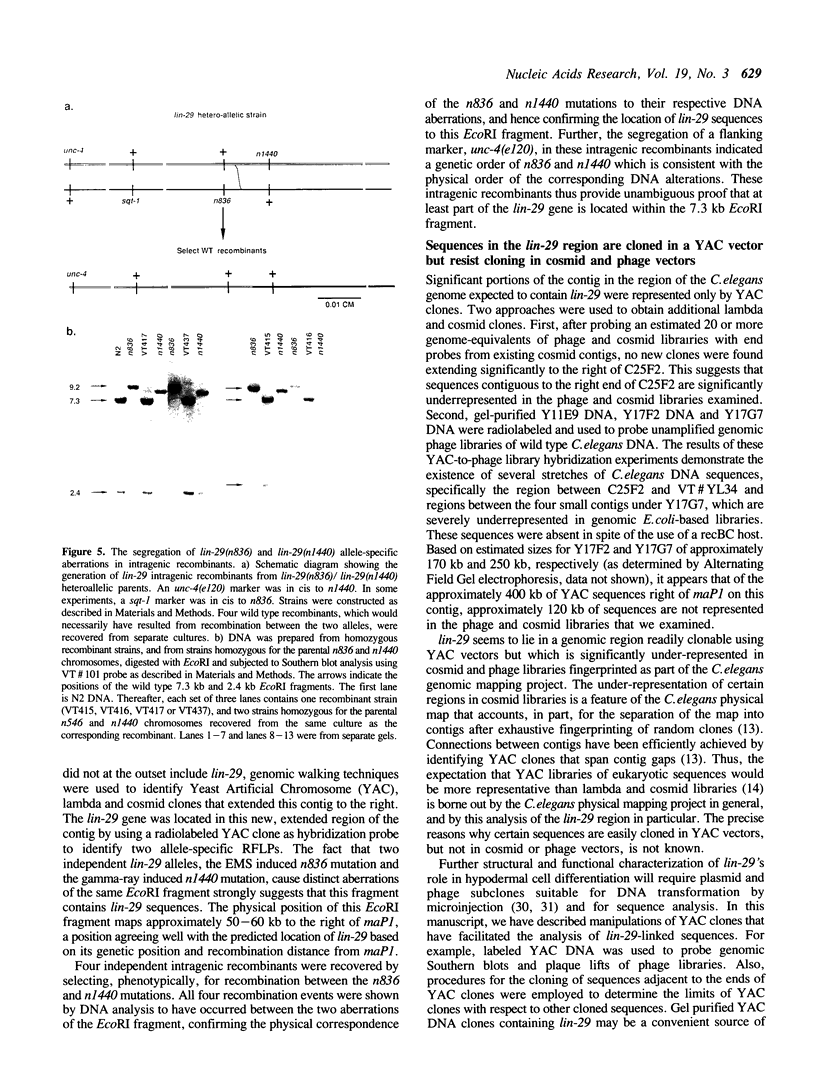

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989 Apr 7;57(1):49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Ambros V., Horvitz H. R. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984 Oct 26;226(4673):409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Ambros V., Horvitz H. R. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1987 Jun;1(4):398–414. doi: 10.1101/gad.1.4.398. [DOI] [PubMed] [Google Scholar]
- Bender W., Spierer P., Hogness D. S. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983 Jul 25;168(1):17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974 May;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. T., Carle G. F., Olson M. V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987 May 15;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson A., Sulston J., Brenner S., Karn J. Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson A., Waterston R., Kiff J., Sulston J., Kohara Y. Genome linking with yeast artificial chromosomes. Nature. 1988 Sep 8;335(6186):184–186. doi: 10.1038/335184a0. [DOI] [PubMed] [Google Scholar]
- Cox G. N., Hirsh D. Stage-specific patterns of collagen gene expression during development of Caenorhabditis elegans. Mol Cell Biol. 1985 Feb;5(2):363–372. doi: 10.1128/mcb.5.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson T. J., Coulson A. R., Sulston J. E., Little P. F. Lorist2, a cosmid with transcriptional terminators insulating vector genes from interference by promoters within the insert: effect on DNA yield and cloned insert frequency. Gene. 1987;53(2-3):275–281. doi: 10.1016/0378-1119(87)90016-3. [DOI] [PubMed] [Google Scholar]
- Herman R. K. Crossover suppressors and balanced recessive lethals in Caenorhabditis elegans. Genetics. 1978 Jan;88(1):49–65. doi: 10.1093/genetics/88.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Matthes H. W., Gait M. J., Brenner S. A new selective phage cloning vector, lambda 2001, with sites for XbaI, BamHI, HindIII, EcoRI, SstI and XhoI. Gene. 1984 Dec;32(1-2):217–224. doi: 10.1016/0378-1119(84)90049-0. [DOI] [PubMed] [Google Scholar]
- Koenig M., Hoffman E. P., Bertelson C. J., Monaco A. P., Feener C., Kunkel L. M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987 Jul 31;50(3):509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Kramer J. M., Johnson J. J., Edgar R. S., Basch C., Roberts S. The sqt-1 gene of C. elegans encodes a collagen critical for organismal morphogenesis. Cell. 1988 Nov 18;55(4):555–565. doi: 10.1016/0092-8674(88)90214-0. [DOI] [PubMed] [Google Scholar]
- Ruvkun G., Ambros V., Coulson A., Waterston R., Sulston J., Horvitz H. R. Molecular genetics of the Caenorhabditis elegans heterochronic gene lin-14. Genetics. 1989 Mar;121(3):501–516. doi: 10.1093/genetics/121.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G., Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989 Mar 23;338(6213):313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- Sager R. Tumor suppressor genes: the puzzle and the promise. Science. 1989 Dec 15;246(4936):1406–1412. doi: 10.1126/science.2574499. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977 Mar;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Wertman K. F., Wyman A. R., Botstein D. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene. 1986;49(2):253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]