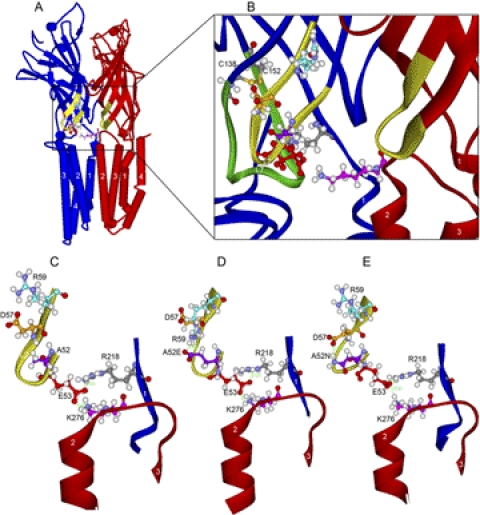
Fig. 6.
Homology models of WT and mutant α1GlyRs based on GLIC. A, WT α1GlyR, a view of two subunits looking along the plane of the membrane from the center of the ion pore. We show two subunits because Lys276 bridges the gap between adjacent subunits (subunit containing Lys276 is colored red) and interacts with the subunit containing Glu53 (colored blue) of the adjacent subunit. B, domain interface in WT α1GlyR. Enlargement of the view in A showing a ribbon rendering of the backbone atoms of loop 2 (yellow portion of both subunits rendered in red and blue ribbons). The residues in the triple salt bridge of Glu53 (red), Arg218 (gray), and Lys276 (pink) are rendered in ball and stick. For reference, loop 7 (Cys-loop) is rendered in green ribbon, and the Cys-loop residues Cys138 and Cys152 are shown (sulfur atoms are orange). C, loop 2 in WT α1GlyR. Further enlargement of WT α1GlyR shown in B is focused on loop 2. The triple salt bridge is established but there are no interactions across the β hairpin of loop2. D, mutant A52E α1GlyR. View of loop 2 in A52E showing the new salt bridge of A52E with R59 and the resulting distortion of the backbone atoms of loop 2. E, mutant A52N α1GlyR. View of loop 2 in A52N showing the new salt bridge of A52N with Asp57 and the resulting distortion of the backbone atoms of loop 2. In the short molecular dynamics simulation, these changes in loop 2 produced small changes in the triple salt bridge of Glu53, Arg218, and Lys276.