Abstract
Background and Aims
The genetics of domestication of yardlong bean [Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis] is of particular interest because the genome of this legume has experienced divergent domestication. Initially, cowpea was domesticated from wild cowpea in Africa; in Asia a vegetable form of cowpea, yardlong bean, subsequently evolved from cowpea. Information on the genetics of domestication-related traits would be useful for yardlong bean and cowpea breeding programmes, as well as comparative genome study among members of the genus Vigna. The objectives of this study were to identify quantitative trait loci (QTLs) for domestication-related traits in yardlong bean and compare them with previously reported QTLs in closely related Vigna.
Methods
Two linkage maps were developed from BC1F1 and F2 populations from the cross between yardlong bean (V. unguiculata ssp. unguiculata cv.-gr. sesquipedalis) accession JP81610 and wild cowpea (V. unguiculata ssp. unguiculata var. spontanea) accession TVnu457. Using these linkage maps, QTLs for 24 domestication-related traits were analysed and mapped. QTLs were detected for traits related to seed, pod, stem and leaf.
Key Results
Most traits were controlled by between one and 11 QTLs. QTLs for domestication-related traits show co-location on several narrow genomic regions on almost all linkage groups (LGs), but especially on LGs 3, 7, 8 and 11. Major QTLs for sizes of seed, pod, stem and leaf were principally located on LG7. Pleiotropy or close linkage of genes for the traits is suggested in these chromosome regions.
Conclusions
This is the first report of QTLs for domestication-related traits in yardlong bean. The results provide a foundation for marker-assisted selection of domestication-related QTLs in yardlong bean and enhance understanding of domestication in the genus Vigna.
Keywords: Cowpea, QTL analysis, domestication, Vigna unguiculata, evolution, yardlong bean
INTRODUCTION
The Leguminosae genus Vigna is a pantropical genus comprising about 100 species mainly found in Africa and Asia (Maréchal et al., 1978). Nine Vigna species have been domesticated, two of which were domesticated in Africa and seven were domesticated in Asia. The African Vigna consists of cowpea [V. unguiculata (L.) Walp.] and bambara groundnut [V. subterranea (L.) Verdc.] (Smartt, 1990). The Asian Vigna comprises mungbean [V. radiata (L.) Wilczek], blackgram [V. mungo (L.) Hepper], moth bean [V. aconitifolia (Jacq.) Maréchal], azuki bean [V. angularis (L.) Ohwi & Ohashi], rice bean [V. umbellata (Thunb.) Ohwi & Ohashi], jungli bean [V. trilobata (L.) Verdc.] and creole bean [V. reflexo-pilosa Hayata] (Tomooka et al., 2002). All domesticated Vigna except creole bean have the chromosome number of 2n = 2x = 22. These crops are adapted to various agroclimatic conditions and fit well into many cropping systems. Dry seeds, young pods and sprouts from these crops are consumed. Plant parts of the crops are used as fodder or hay for farm animals. Among these Vigna crops, cowpea is the most important in terms of planting area.
Yardlong bean [Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis] is characterized by its very long (30–90 cm in length) pods with seeds usually 8–12 mm long. It is also known as asparagus bean, string bean, snake bean and sitao. Unlike other Vigna crops, which are grown primarily for seeds, yardlong bean is cultivated mainly for crisp and tender pods that are consumed both fresh and cooked. Yardlong bean is believed to have been domesticated from cultivated cowpea in Asia. It is widely grown as a vegetable in China, and South and South-East Asia.
Yardlong bean and cowpea differ phenotypically as a result of divergent selection during the evolution of the crop. The domestication syndrome is a suite of morphological and physiological traits that distinguish domesticated crops from their wild ancestors (Hammer, 1984). Domestication-related traits include changes in plant architecture (e.g. no branching in maize, determinate growth habit in beans), gigantism in the consumed plant organs (e.g. seed size in bean), reduced seed dispersal (i.e. non-shattering or non-dehiscence in bean and rice) and loss of seed dormancy. Crop domestication is an accelerated evolutionary process that is the result of the synergistic effect of human (both intentional and unintentional selection) and natural selection. Plant scientists are interested in studying the genetic basis of crop domestication with an ultimate goal of identifying useful allele(s), gene(s) or genome region(s) that have not been exploited in the wild relatives of the cultivated crops and could have a positive impact on crop improvement. The search and localization for genes involved in crop domestication are performed by means of quantitative trait loci (QTL) analyses in many crops. Some genes for domestication have been cloned (Purugganan and Fuller, 2009; Izawa et al., 2009). In addition, divergence within the primary gene pool of cowpea to a crop for seeds and pods outside the natural range of the wild relatives of cowpea can provide insight into crop evolution and agriculture. Unfortunately, there is a lack of archaeobotanical information on cowpea and particularly yardlong bean in Asia (but see Fuller and Harvey, 2006).
The genetics of domestication syndrome traits have been reported in a limited number of legume crops, including azuki bean, common bean, pea, soybean and rice bean. Among the nine cultivated Vigna species, comprehensive QTL mappings for domestication syndrome traits were reported for azuki bean (Kaga et al., 2008) and rice bean (Isemura et al., 2010), both of which are Asian Vigna crops. QTL analyses for domestication traits in cowpea and yardlong bean have recently been published (Andargie et al., 2011; Xu et al., 2011). However, these papers examined only two traits of domestication-related characters.
The genetics of domestication of yardlong bean is of particular interest because the genome of the bean has experienced divergent domestication from cowpea grown primarily for its seeds. Cowpea was domesticated from wild cowpea in Africa; in Asia, cultivated cowpea or its weedy relative was subsequently selected for the vegetable crop, yardlong bean. Information on the genetics of domestication-related traits would be useful for yardlong bean and cowpea breeding programmes, and for comparative genome studies among members of the genus Vigna.
The objectives of this study were to identify QTLs for domestication-related traits in yardlong bean and to compare them with previously reported QTLs in closely related Vigna species.
MATERIALS AND METHODS
Mapping populations
Mapping populations comprised F2 and BC1F1 populations. They were derived from a cross between yardlong bean accession ‘JP81610’ and wild cowpea (V. unguiculata ssp. unguiculata var. spontanea) accession ‘JP89083 = TVnu457’. JP81610 is a landrace from Sri Lanka, whereas JP89083 originated from Africa. Both accessions were obtained from the Gene Bank, National Institute of Agrobiological Sciences (NIAS), Tsukuba, Japan. Pollen of JP89083 was pollinated onto JP81610 to produce F1 hybrid plants. An F1 plant was self-pollinated to produce the F2 population and, at the same time, another F1 plant was crossed as female parent with JP81610 to develop the BC1F1 population. The F2 population consisted of 188 plants, while the BC1F1 population consisted of 190 plants. The BC1F1 population was the same as that reported for linkage map construction by Kongjaimun et al. (2012).
DNA extraction
Total genomic DNA of parents and F2 and BC1F1 populations were extracted from fresh leaf tissue using the CTAB method (Lodhi et al., 1994). DNA concentration was estimated and adjusted to 5 ng μL−1 for simple sequence repeat (SSR) analyses by comparing with a known concentration of standard λ-DNA on 1·5 % agarose gel.
Trait measurement
Twenty-four traits related to fitness and domestication were evaluated following Kaga et al. (2008) (Table 1). Of these, 21 were treated as quantitative traits and three, pod dehiscence, epicotyl colour and seed coat colour, were treated as qualitative traits. The F2 population of 188 plants, together with ten plants of each parent, were grown in a net house at the experimental field of Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom, Thailand (13°48′N, 99°5′E) from October 2008 to February 2009. The BC1F1 population of 190 plants, together with ten plants of each parent, were grown in 20-cm-diameter pots in a vinyl greenhouse of the Gene Bank, National Institute of Agrobiological Sciences (NIAS), Tsukuba, Japan (36°2′N, 140°8′E) from May to August 2009.
Table 1.
Domestication-related traits examined in F2 and BC1F1 populations of the cross between yardlong bean and wild cowpea (based on Kaga et al., 2008: table 1)
| General attribute | Organ | Trait | Trait abbreviation | QTL/gene | Evaluation method | Evaluated population |
|---|---|---|---|---|---|---|
| Seed dormancy | Seed | Seed coat permeability (%) | SDP | Sdp | Percentage of imbibed seeds at 25 °C in incubator (use 20 seeds) | BC1F1:2/F2:3 |
| Pod dehiscence | Pod | Number of twists (count) | PDT | pdt | Number of twists along the length of the shattered pod | BC1F1 |
| Pod dehiscence | PDD | Pdd | Dehiscence or indehiscence | F2 | ||
| Gigantism | Seed | 100-seed weight (g) | SD100WT | Sd100wt | Weight of 100 seeds (use 100 seeds) | BC1F1:2/F2:3 |
| Length (mm) | SDL | Sdl | Maximum distance from top to bottom of the seed (use 10 seeds) | BC1F1:2/F2:3 | ||
| Width (mm) | SDW | Sdw | Maximum distance from hilum to its opposite side (use 10 seeds) | BC1F1:2/F2:3 | ||
| Thickness (mm) | SDT | Sdt | Maximum distance between both sides of the hilum (use 10 seeds) | BC1F1:2/F2:3 | ||
| Pod | Length (cm) | PDL | Pdl | Length of straight pod (use 10 pods) | BC1F1/F2 | |
| Width (mm) | PDW | Pdw | Maximum width (use 10 pods) measured after soaking in water to flatten the pod to measure | BC1F1/F2 | ||
| Spacing between seeds (mm) | PDSBS* | Pdsbs | Spacing between seeds is calculated by formula: [(PDL*10) – (SDL*SDNPPD)]/SDNPPD | BC1F1/F2 | ||
| Stem | Thickness (mm) | STT | Stt | Stem diameter under the primary leaf (measured at flowering stage) | BC1F1/F2 | |
| Leaf | Primary leaf length (cm) | LFPL | Lfpl | Distance from pulvinus to leaf tip | BC1F1/F2 | |
| Primary leaf width (cm) | LFPW | Lfpw | Maximum width | BC1F1/F2 | ||
| Plant type | Epicotyl | Length (cm) | ECL | Ecl | Length from cotyledon to primary leaf | BC1F1/F2 |
| Stem | Length (up to 10th node) (cm) | STL10 | Stl10 | Length from node on primary leaf to node 10 of trifoliate leaf | BC1F1/F2 | |
| Length (whole) | STLW* | Stlw | Length from node on primary leaf to terminal shoot | BC1F1 | ||
| Branch | Number (count) | BRN | Brn | Number of branches on main stem from node 1 to node 10 of trifoliate leaf (measured at post maturity stage just before discarding) | BC1F1/F2 | |
| Earliness | Flower | Days to first flower (day) | FLD | Fld | Number of days from planting to 1st flowering | BC1F1/F2 |
| Pod | Days to maturity of 1st pod (day) | PDDM | Pddm | Number of days from 1st flowering to harvesting of 1st pod | BC1F1/F2 | |
| Yield potential | Seed | Total weight (g) | SDTWT | Sdtwt | Total weight of harvested seeds | BC1F1/F2 |
| Number of seeds per pod (seeds/pod) | SDNPPD | Sdnppd | Number of seeds per pod | BC1F1/F2 | ||
| Pod | Total number (pod) | PDTN | Pdtn | Total number of harvested pods | BC1F1/F2 | |
| Pigmentation | Epicotyl | Epicotyl colour | ECC | Ecc | Red or green | BC1F1/F2 |
| Seed | Seed coat colour | SDC | Sdc | Black or brown | BC1F1/F2 |
* Not included in Kaga et al. (2008).
In both F2 and BC1F1 populations, seedling traits, i.e. primary leaf length (LFPL), primary leaf width (LFPW), epicotyl colour (ECC) and epicotyl length (ECL), were recorded when the first trifoliate leaf opened. Stem length (STL), stem thickness (STT) and branch number (BRN) were recorded when the tenth trifoliate leaf was fully developed. Whole stem length (STLW) was an additional trait evaluated only in the BC1F1 population (Table 1).
Seed-related traits were investigated using seeds and pods from both F2 and BC1F1 plants. Seed coat permeability (SDP) as an index of seed dormancy was determined using ten unscarified seeds from each BC1F1 plant. The seeds were placed on filter paper, incubated at 25 °C for 7 d, and the number of seeds that imbibed water was counted daily. Seed dimensions, namely seed length (SDL), seed width (SDW) and seed thickness (SDT), were the averages of ten seeds. The 100-seed weight (SD100WT) was evaluated using intact seeds of each plant.
Pod traits, i.e. pod length (PDL), pod width (PDW), pod dehiscence (PDD) and number of twists along the length of dehisced pods (PDT) when kept at room temperature, were evaluated from ten pods of each plant. PDT was used as an index of pod structure. Pod dehiscence was scored as dehiscent or indehiscent on the basis of whether seeds shattered from pods or not.
The number of days from planting to first flowering (FLD) was recorded. Days to first mature pod (PDDM) was defined as the number of days from first flowering to harvesting of the first mature pod. After harvesting all pods, total pod number (PDTN) and total seed weight (SDTWT) were measured in each plant. The number of seeds per pod (SDNPPD) was recorded using ten pods. Spacing between seeds (PDSBS) was evaluated with the formula: [(PDL*10) – (SDL*SDNPPD)]/SDNPPD using the data from ten pods. Seed colour from each plant was scored as black or brown.
SSR marker analysis
A genetic linkage map based on the F2 population was constructed using framework SSR markers that were distributed throughout the BC1F1 genetic linkage map (Kongjaimun et al., 2012). In total, 113 SSR markers, located at about 10-cM intervals on each linkage group of the BC1F1 genetic linkage map, were chosen to genotype the F2 population. The polymorphic SSR markers consisted of 78 from cowpea, 26 from azuki bean and nine from mungbean.
The PCR reaction mixture and multiplex PCR were the same as those described by Kongjaimun et al. (2012). The PCR thermal cycling programmes for azuki bean, cowpea and mungbean SSRs were set as described by Wang et al. (2004). After amplification, allele size(s) of each primer pair was determined according to Kongjaimun et al. (2012).
Data analysis
Mean, standard deviation and broad-sense heritability were calculated, and the frequency distribution of phenotypes in the F2 and BC1F1 populations were examined for each trait. The correlation coefficient (r) between traits was also calculated. The total number of seeds per plant was used as an index of seed productivity and treated as the dependent (y) variable. As stem length, branch number, leaf size (the product of maximum length and width), 100-seed weight, pod length, total number of pods per plant and number of seeds per pod are possibly correlated to the total number of seeds, these traits were treated as the independent (x) variables.
Linkage map construction and QTL analysis
The linkage map of the F2 population was constructed by JoinMap 4·0 (Van Ooijen, 2006). Chi-square analysis was performed for the goodness-of-fit to 1 : 2 : 1 segregation ratio of each marker at a probability (P) of 0·05, 0·01 and 0·001. A minimum LOD score of 3 and recombination frequency of 0·25 in the Kosambi mapping function (Kosambi, 1944) were used. Double crossovers between adjacent loci were confirmed manually. QTL analysis was conducted using the software package MultiQTL ver. 2·6 according to the procedures described by Kaga et al. (2008). The QTLs were named after Somta et al. (2006). Randomness of the genomic distribution of the QTLs was determined using chi-square tests (Isemura et al., 2007). Randomness of the QTLs along a linkage group was tested using a Poisson distribution test (Somta et al., 2006; Isemura et al., 2007).
RESULTS
Linkage map construction
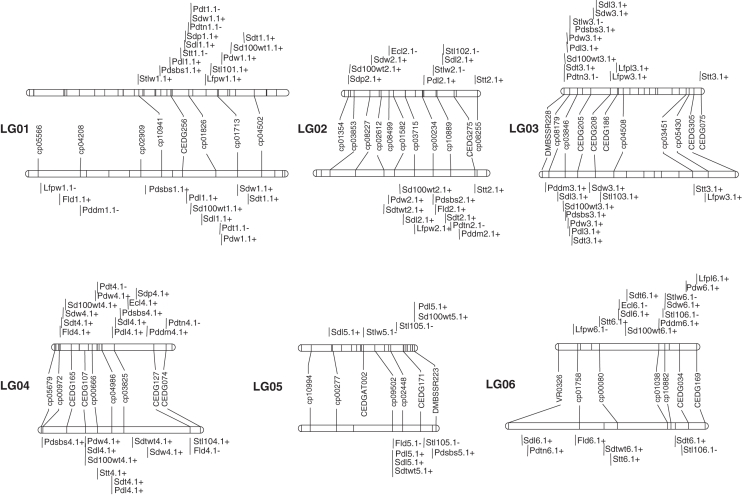
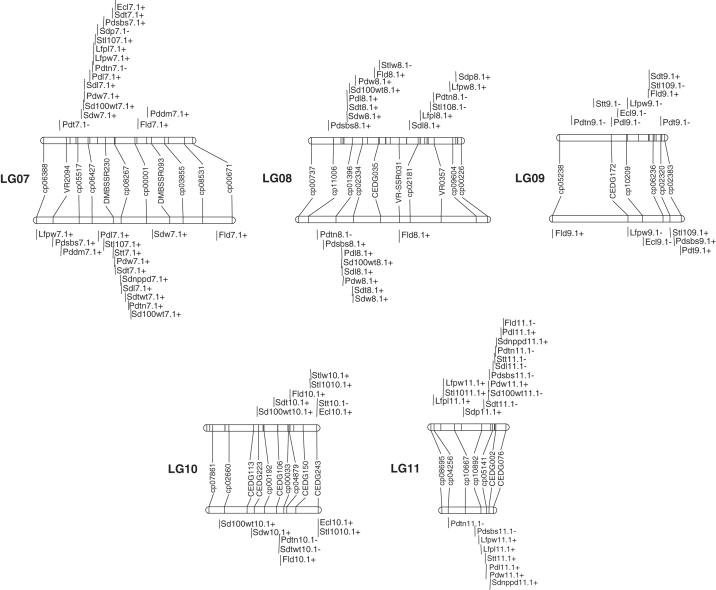
The BC1F1 map constructed by Kongjaimun et al. (2012) across 11 linkage groups (LGs) was used. The map consists of 226 SSR markers (165 from cowpea, 46 from azuki bean and 15 from mungbean) and spans a total distance of 852·4 cM. The F2 map constructed here using 113 SSR markers (78 from cowpea, 26 from azuki bean and nine from mungbean) covering 977·1 cM with an average distance between the adjacent markers of 9·58 cM (Table 2). The length of the LGs ranged from 28·3 cM (LG11) to 140·4 cM (LG01) with a mean of 88·83 cM. The number of marker loci per LG varied from six (LG9) to 16 (LG1) with a mean of 10·27. The length of the F2 map was equivalent to 114·6 % of the BC1F1 map. All of the LGs except LG11 had a length of 60 cM or longer. The largest gap of 41·3 cM was on LG9. Segregation distortion was observed at 48·7 % (55 of 113) of mapped markers (P < 0·05). The distorted markers were distributed on all LGs (Fig. 1). In most cases, on the same LG the distorted markers appeared to be clustered or close to one another. All the markers on LG11 and nine out of 11 (81·8 %) markers on LG7 showed distortion (Supplementary Data, Table S1). Orders and linkages of all the markers were the same in both linkage maps.
Table 2.
Number of markers and average distance between markers in each linkage group in the yardlong bean map estimated from the F2 population
| No. of SSR markers |
||||||
|---|---|---|---|---|---|---|
| Linkage group | Length (cM) | Average interval (cM) | Total | Cowpea | Azuki bean | Mung bean |
| 1 | 140·4 | 9·36 | 16 | 12 | 4 | 0 |
| 2 | 93·4 | 8·49 | 12 | 9 | 3 | 0 |
| 3 | 105·7 | 8·13 | 14 | 10 | 3 | 1 |
| 4 | 87·2 | 10·9 | 9 | 5 | 4 | 0 |
| 5 | 73·7 | 9·21 | 9 | 6 | 1 | 2 |
| 6 | 107·7 | 13·46 | 9 | 6 | 2 | 1 |
| 7 | 107·1 | 10·71 | 11 | 8 | 0 | 3 |
| 8 | 103·4 | 11·49 | 10 | 7 | 1 | 2 |
| 9 | 70·5 | 14·09 | 6 | 5 | 1 | 0 |
| 10 | 59·8 | 6·64 | 10 | 5 | 5 | 0 |
| 11 | 28·3 | 4·72 | 7 | 5 | 2 | 0 |
| Total | 977·1 | 9·58 | 113 | 78 | 26 | 9 |
Fig. 1.
A genetic linkage map of yardlong bean based on SSR markers. This map was constructed from 188 F2 individuals of V. unguiculata ssp. unguiculata cv.-gr. sesquipedalis × V. unguiculata ssp. unguiculata var. spontanea. Map distances (cM) are shown on the left side and marker names on the right side of the linkage groups. ‘cp-’ and ‘CED-’ represent the SSR marker loci from cowpea and azuki bean, respectively, and ‘DMBSSR-’, ‘VR-’ and ‘VR-SSR-’ represent the SSR marker loci from mungbean. Asterisks indicate significant segregation distortion at the *5, **1 and ***0·1 % significance levels, respectively.
Variation of domestication-related traits
The mean, range and standard deviation of traits in the parents, and F2 and BC1F1 populations as well as their broad-sense heritabilities are shown in Tables 3 and 4. Means of the F2 population generally fell between those of the parents for all traits except days to maturity of first pod and number of seeds per pod. Similarly, means of the BC1F1 population were between their parents for all traits except days to first flowering and number of seeds per pod. The parents were clearly different in all traits observed. The cultivated parent showed higher values than the wild parent for seed coat permeability, number of twists along the pod, stem length (1st to 10th nodes and the 1st node to terminal shoot) and the size of organs such as leaf, stem, seed and pod. A pronounced difference was observed for SD100WT, PDL, PDW, PDSBS, STL10 and PDTN. Regarding qualitative traits, the cultivated parent had a black seed coat and indehiscent pod whereas the wild parent had a black mottled over tan seed coat and pods were dehiscent. Both parents had green epicotyls.
Table 3.
Mean, standard deviation, minimum, maximum and heritability values of parents, and F2 or F2:3 populations derived from the cross between cultivated yardlong bean and wild cowpea
| Cultivated yardlong bean, JP81610 |
Wild cowpea, TVnu457 |
F2 or F2:3 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Units | Mean | s.d. | Min. | Max. | Mean | s.d. | Min. | Max. | Mean | s.d. | Min. | Max. | Heritability (%) |
| SDP | (%) | 100·0 | 0·0 | 100·0 | 100·0 | 0·0 | 0·0 | 0·0 | 0·0 | 78·2 | 36·5 | 0·0 | 100·0 | 100·0 |
| PDD | – | Indehiscence | Dehiscence | Dehiscence : indehiscence* | ||||||||||
| SD100WT | g | 19·4 | 1·4 | 16·6 | 20·9 | 2·5 | 0·2 | 2·2 | 2·7 | 5·7 | 1·9 | 2·0 | 16·1 | 72·6 |
| SDL | mm | 11·9 | 0·4 | 11·3 | 12·5 | 3·9 | 0·2 | 3·6 | 4·1 | 7·2 | 1·1 | 4·3 | 9·9 | 92·5 |
| SDW | mm | 3·7 | 0·2 | 3·3 | 4·0 | 2·0 | 0·1 | 1·8 | 2·2 | 2·7 | 0·3 | 0·8 | 3·8 | 81·0 |
| SDT | mm | 6·2 | 0·2 | 5·9 | 6·5 | 2·7 | 0·3 | 2·4 | 3·9 | 4·0 | 0·5 | 2·6 | 5·8 | 65·7 |
| PDL | cm | 78·1 | 3·1 | 72·9 | 84·0 | 9·9 | 0·4 | 8·8 | 10·4 | 21·3 | 5·8 | 10·2 | 41·3 | 85·7 |
| PDW | mm | 11·5 | 0·8 | 10·0 | 13·0 | 0·5 | 0·0 | 0·4 | 0·5 | 0·7 | 0·1 | 0·5 | 1·0 | 49·9 |
| PDSBS | mm | 26·6 | 1·8 | 23·8 | 30·8 | 1·9 | 0·2 | 1·6 | 2·2 | 9·9 | 3·7 | 2·1 | 26·3 | 87·8 |
| STT | mm | 15·3 | 2·0 | 11·2 | 18·4 | 9·7 | 1·8 | 6·3 | 11·9 | 13·4 | 2·0 | 7·4 | 18·8 | 15·1 |
| LFPL | cm | 7·6 | 0·7 | 6·5 | 8·8 | 2·9 | 0·4 | 2·2 | 3·4 | 4·7 | 0·6 | 2·4 | 6·1 | 18·0 |
| LFPW | cm | 5·2 | 0·5 | 4·5 | 6·0 | 1·6 | 0·2 | 1·2 | 2·0 | 2·9 | 0·4 | 1·9 | 3·7 | 2·6 |
| ECL | cm | 6·6 | 0·9 | 5·0 | 8·0 | 4·2 | 0·5 | 3·2 | 5·0 | 5·2 | 1·1 | 2·5 | 8·5 | 62·2 |
| STL10 | cm | 120·8 | 9·5 | 105·0 | 134·3 | 30·0 | 9·5 | 16·0 | 46·0 | 68·3 | 19·9 | 30·0 | 118·5 | 77·1 |
| FLD | day | 55·1 | 3·5 | 50·0 | 62·0 | 43·9 | 5·9 | 37·0 | 62·0 | 43·9 | 4·3 | 37·0 | 58·0 | 0 |
| PDDM | day | 18·6 | 4·1 | 15·0 | 28·0 | 14·6 | 3·3 | 9·0 | 19·0 | 20·6 | 2·2 | 15·0 | 29·0 | 0 |
| SDTWT | g | 209·0 | 55·5 | 115·0 | 307·5 | 94·9 | 23·3 | 54·3 | 131·8 | 161·1 | 99·8 | 12·9 | 631·1 | 81·8 |
| SDNPPD | count | 20·3 | 0·7 | 19·1 | 21·6 | 17·3 | 0·6 | 15·9 | 17·9 | 12·8 | 3·0 | 7·0 | 20·3 | 95·7 |
| PDTN | count | 128·5 | 20·3 | 59·0 | 130·0 | 504·8 | 131·9 | 248·0 | 701·0 | 461·9 | 248·6 | 15·0 | 1609·0 | 85·6 |
| ECC | – | Green | Green | Green | ||||||||||
| SDC | – | Black | Brown | Brown : black† | ||||||||||
* = 111:77 (ratio = 9 : 7, χ2 = 0·60, P = 0·44).
† = 33:155 (ratio = 3 : 13, χ2 = 0·18, P = 0·67).
Table 4.
The mean, standard deviation, minimum, maximum and heritability values of parents, and the BC1F1 or BC1F1:2 populations derived from the cross between cultivated yardlong bean and wild cowpea
| Cultivated yardlong bean, JP81610 |
Wild cowpea, TVnu457 |
BC1F1 or BC1F1:2 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Units | Mean | s.d. | Min. | Max. | Mean | s.d. | Min. | Max. | Mean | s.d. | Min. | Max. | Heritability (%) |
| SDP | (%) | 97·7 | 2·7 | 93·0 | 100·0 | 0·0 | 0·0 | 0·0 | 0·0 | 46·9 | 31·4 | 0·0 | 100·0 | 99·6 |
| PDT | count | 0·0 | 0·0 | 0·0 | 0·0 | 6·2 | 0·3 | 5·8 | 6·6 | 3·4 | 4·6 | 0·0 | 13·0 | 99·9 |
| SD100WT | g | 17·5 | 1·4 | 14·4 | 19·6 | 2·4 | 0·2 | 1·8 | 2·6 | 10·5 | 1·9 | 6·4 | 18·7 | 71·4 |
| SDL | mm | 11·2 | 0·4 | 10·5 | 11·9 | 3·6 | 0·1 | 3·5 | 3·9 | 8·9 | 0·8 | 4·6 | 10·7 | 85·2 |
| SDW | mm | 6·3 | 0·2 | 5·9 | 6·7 | 2·5 | 0·1 | 2·3 | 2·6 | 5·0 | 0·4 | 3·9 | 5·9 | 78·1 |
| SDT | mm | 4·1 | 0·2 | 3·9 | 4·4 | 1·8 | 0·1 | 1·7 | 1·9 | 3·4 | 0·2 | 2·7 | 4·1 | 72·1 |
| PDL | cm | 64·4 | 5·7 | 56·8 | 73·9 | 8·5 | 0·3 | 8·0 | 9·0 | 31·6 | 7·8 | 15·1 | 57·1 | 73·1 |
| PDW | mm | 1·3 | 0·1 | 1·2 | 1·4 | 0·5 | 0·0 | 0·5 | 0·5 | 0·9 | 0·1 | 0·7 | 1·3 | 88·2 |
| PDSBS | mm | 30·0 | 3·5 | 25·1 | 38·1 | 3·1 | 0·7 | 2·1 | 4·0 | 17·4 | 6·7 | 4·4 | 40·2 | 86·1 |
| STT | mm | 7·4 | 1·1 | 5·7 | 9·1 | 5·4 | 1·0 | 4·2 | 6·8 | 6·0 | 1·1 | 3·9 | 8·5 | 0 |
| LFPL | cm | 8·0 | 0·7 | 7·2 | 9·3 | 3·4 | 0·3 | 2·9 | 4·0 | 5·9 | 0·5 | 4·3 | 7·5 | 6·3 |
| LFPW | cm | 5·5 | 0·3 | 5·0 | 5·9 | 1·8 | 0·3 | 1·2 | 2·2 | 3·7 | 0·3 | 2·8 | 4·5 | 13·9 |
| ECL | cm | 4·2 | 0·4 | 3·6 | 5·0 | 2·2 | 0·4 | 1·7 | 3·0 | 3·3 | 0·6 | 2·0 | 5·7 | 65·8 |
| STL10 | cm | 177·8 | 14·1 | 157·0 | 195·5 | 63·6 | 6·3 | 53·0 | 73·0 | 142·4 | 32·9 | 61·0 | 219·5 | 89·0 |
| STLW | cm | 154·8 | 23·8 | 121·0 | 195·0 | 21·6 | 14·6 | 11·0 | 48·0 | 99·5 | 45·4 | 17·0 | 220·0 | 81·1 |
| BRN | count | 2·7 | 1·6 | 1·0 | 6·0 | 7·1 | 0·9 | 6·0 | 9·0 | 3·8 | 1·8 | 0·0 | 9·0 | 45·8 |
| FLD | day | 57·6 | 2·7 | 55·0 | 64·0 | 56·8 | 4·4 | 50·0 | 66·0 | 56·0 | 4·4 | 47·0 | 67·0 | 29·5 |
| PDDM | day | 18·4 | 1·5 | 16·0 | 21·0 | 12·3 | 0·9 | 11·0 | 14·0 | 15·9 | 1·7 | 7·0 | 24·0 | 47·2 |
| SDTWT | g | 15·4 | 5·1 | 10·9 | 28·0 | 12·8 | 4·2 | 7·0 | 22·4 | 15·7 | 5·5 | 4·6 | 31·5 | 27·8 |
| SDNPPD | count | 15·7 | 2·2 | 11·6 | 19·8 | 12·9 | 1·1 | 11·3 | 14·3 | 12·5 | 3·2 | 5·1 | 19·5 | 69·7 |
| PDTN | count | 6·7 | 2·2 | 3·0 | 10·0 | 89·3 | 21·6 | 47·0 | 119·0 | 17·3 | 8·1 | 2·0 | 50·0 | 0 |
| ECC | – | Green | Green | Green | ||||||||||
| SDC | – | Black | Brown | Black | ||||||||||
The measured traits showed nearly a normal distribution in both populations (Supplementary Data, Figs S1 and S2). The mean percentage of imbibed seeds (SDP) for the F2 population showed a biased distribution towards the cultivated parent. Clear transgressive segregation was observed in STT, PDSBS, ECL, STL10, STLW, BRN, FLD, PDDM, SDTWT, SDNPPD and PDTN in both the F2 and the BC1F1 population.
Generally, the traits measured showed high broad-sense heritability (Tables 3 and 4). SDP, SDTWT, SDL, SDW, PDL, PDSBS and STL10 showed high heritability (>70 %) whereas STT, LFPL, LFPW, ECL, FLD and PDDM showed medium to low heritability (<70 %). SDT and PDW showed high heritability in only the BC1F1 population, while SDTWT, SDNPPD and PDTN showed high heritability in only the F2 population.
There were significant positive correlations (P < 0·05) between related traits, such as between pod length and pod width, and between seed size-related traits and pod size-related traits (Supplementary Data, Tables S2 and S3). PDT and PDTN were negatively correlated with seed-related traits, STLW and earliness traits (FLD and PDDM).
QTLs for domestication-related traits
The results of the QTL analysis for each trait in each population are shown in Table 5. Only QTLs with P ≤ 0·05 were considered. In total, 153 QTLs were identified for the 21 traits. One to 11 QTLs were detected for each trait except for BRN and SDTWT in the BC1F1 population, from which no significant QTL was detected.
Table 5.
QTLs detected in the BC1F1:2, F2 and F2:3 populations derived from the cross between cultivated yardlong bean and wild cowpea
|
BC1F1:2 |
F2:3 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | QTL name | LG | LOD | P | Loci (cM) | PVE (%) | Additive effect | LG | LOD | P | Loci (cM) | PVE (%) | Additive effect | Dominant effect |
| SDP | Sdp1·1 + | 1 | 27·3 | 0·0009 | 90·2 | 41·1 | 8·1 | 1 | ||||||
| Sdp2·1 + | 2 | 4·3 | 0·0009 | 2·2 | 4·3 | 2·6 | 2 | |||||||
| Sdp4·1 + | 4 | 2·2 | 0·0112 | 46·9 | 2·9 | 2·1 | 4 | NI | ||||||
| Sdp7·1– | 7 | 1·9 | 0·0300 | 30·8 | 1·9 | –1·7 | 7 | |||||||
| Sdp8·1 + | 8 | 3·1 | 0·0019 | 82·8 | 3·0 | 2·2 | 8 | |||||||
| Sdp11·1 + | 11 | 4·9 | 0·0009 | 17·5 | 5·2 | 2·9 | 11 | |||||||
| PDT | Pdt1·1– | 1 | 11·7 | 0·0009 | 93·4 | 11·8 | –3·3 | 1 | NI | |||||
| Pdt4·1– | 4 | 3·0 | 0·0009 | 24·5 | 2·6 | –1·5 | 4 | |||||||
| Pdt7·1– | 7 | 30·1 | 0·0009 | 9·3 | 47·6 | –6·5 | 7 | |||||||
| Pdt9·1– | 9 | 2·2 | 0·0084 | 58·1 | 1·9 | –1·3 | 9 | |||||||
| SD100WT | Sd100wt1·1 + | 1 | 17·2 | 0·0009 | 116·2 | 11·0 | 1·3 | 1 | 18·9 | 0·0009 | 89·0 | 16·8 | 1·1 | –0·5 |
| Sd100wt2·1 + | 2 | 8·4 | 0·0009 | 2·9 | 4·6 | 0·8 | 2 | 4·7 | 0·0009 | 45·7 | 3·6 | 0·5 | 0·3 | |
| Sd100wt3·1 + | 3 | 12·4 | 0·0009 | 0·6 | 7·1 | 1·0 | 3 | 16·4 | 0·0009 | 11·5 | 18·7 | 1·2 | 0·1 | |
| Sd100wt4·1 + | 4 | 10·6 | 0·0009 | 6·2 | 6·1 | 1·0 | 4 | 12·8 | 0·0009 | 24·3 | 8·9 | 0·7 | 0·4 | |
| Sd100wt5·1 + | 5 | 5·7 | 0·0009 | 64·9 | 2·9 | 0·7 | 5 | |||||||
| Sd100wt6·1 + | 6 | 5·1 | 0·0009 | 38·7 | 5·2 | 0·9 | 6 | |||||||
| Sd100wt7·1 + | 7 | 31·2 | 0·0009 | 22·9 | 24·6 | 1·9 | 7 | 11·9 | 0·0009 | 51·6 | 13·1 | 1·0 | 0·3 | |
| Sd100wt8·1 + | 8 | 9·5 | 0·0009 | 21·6 | 5·5 | 0·9 | 8 | 14·1 | 0·0009 | 23·7 | 11·4 | 0·9 | 0·0 | |
| Sd100wt10·1 + | 10 | 8·0 | 0·0009 | 28·9 | 4·3 | 0·8 | 10 | 4·4 | 0·0009 | 5·8 | 2·6 | 0·4 | 0·2 | |
| Sd100wt11·1– | 11 | 11·8 | 0·0009 | 31·9 | 7·6 | –1·1 | 11 | |||||||
| SDL | Sdl1·1 + | 1 | 12·6 | 0·0009 | 89·8 | 10·4 | 0·5 | 1 | 29·4 | 0·0009 | 94·9 | 20·8 | 0·7 | –0·1 |
| Sdl2·1 + | 2 | 6·4 | 0·0009 | 56·5 | 4·8 | 0·4 | 2 | 8·0 | 0·0009 | 47·7 | 2·8 | 0·2 | 0·2 | |
| Sdl3·1 + | 3 | 8·8 | 0·0009 | 18·5 | 7·2 | 0·4 | 3 | 25·0 | 0·0009 | 8·6 | 18·8 | 0·7 | 0·3 | |
| Sdl4·1 + | 4 | 16·0 | 0·0009 | 35·0 | 13·5 | 0·6 | 4 | 21·9 | 0·0009 | 24·3 | 7·5 | 0·4 | 0·1 | |
| Sdl5·1 + | 5 | 3·8 | 0·0009 | 15·9 | 2·7 | 0·3 | 5 | 5·9 | 0·0009 | 49·9 | 1·6 | 0·2 | 0·0 | |
| Sdl6·1 + | 6 | 2·4 | 0·0075 | 34·5 | 2·1 | 0·2 | 6 | 4·9 | 0·0009 | 7·3 | 1·7 | 0·2 | 0·2 | |
| Sdl7·1 + | 7 | 13·6 | 0·0009 | 23·4 | 13·5 | 0·6 | 7 | 41·8 | 0·0009 | 46·3 | 26·5 | 0·7 | 0·5 | |
| Sdl8·1 + | 8 | 12·7 | 0·0009 | 56·2 | 9·8 | 0·5 | 8 | 22·6 | 0·0009 | 23·9 | 9·1 | 0·5 | 0·0 | |
| Sdl11·1– | 11 | 11·9 | 0·0009 | 34·4 | 9·3 | –0·5 | 11 | |||||||
| SDW | Sdw1·1 + | 1 | 22·1 | 0·0009 | 93·2 | 18·4 | 0·3 | 1 | 3·2 | 0·0047 | 114·5 | 4·1 | 0·1 | 0·0 |
| Sdw2·1 + | 2 | 11·1 | 0·0009 | 15·6 | 8·2 | 0·2 | 2 | |||||||
| Sdw3·1 + | 3 | 19·6 | 0·0009 | 16·8 | 17·2 | 0·3 | 3 | 3·1 | 0·0094 | 25·8 | 4·2 | 0·1 | 0·1 | |
| Sdw4·1 + | 4 | 4·8 | 0·0009 | 4·7 | 3·3 | 0·1 | 4 | 6·6 | 0·0009 | 58·3 | 11·7 | 0·2 | 0·0 | |
| Sdw6·1 + | 6 | 2·5 | 0·0075 | 61·2 | 1·6 | 0·1 | 6 | |||||||
| Sdw7·1 + | 7 | 22·9 | 0·0009 | 21·6 | 20·9 | 0·4 | 7 | 5·3 | 0·0009 | 63·4 | 18·5 | 0·2 | 0·2 | |
| Sdw8·1 + | 8 | 6·7 | 0·0009 | 20·3 | 4·6 | 0·2 | 8 | 8·2 | 0·0009 | 29·9 | 8·8 | 0·1 | 0·0 | |
| Sdw10·1 + | 10 | 10 | 8·7 | 0·0009 | 23·3 | 10·0 | 0·2 | 0·0 | ||||||
| SDT | Sdt1·1 + | 1 | 6·8 | 0·0009 | 123·9 | 6·4 | 0·1 | 1 | 12·6 | 0·0009 | 120·3 | 11·3 | 0·2 | –0·1 |
| Sdt2·1 + | 2 | 2 | 9·0 | 0·0009 | 70·7 | 7·5 | 0·2 | 0·1 | ||||||
| Sdt3·1 + | 3 | 11·8 | 0·0009 | 0·0 | 10·5 | 0·2 | 3 | 21·4 | 0·0009 | 16·4 | 26·4 | 0·3 | 0·1 | |
| Sdt4·1 + | 4 | 8·7 | 0·0009 | 3·8 | 7·7 | 0·1 | 4 | 5·5 | 0·0009 | 38·7 | 3·8 | 0·1 | 0·0 | |
| Sdt6·1 + | 6 | 4·2 | 0·0009 | 39·4 | 5·3 | 0·1 | 6 | 5·6 | 0·0009 | 91·4 | 3·9 | 0·1 | 0·0 | |
| Sdt7·1 + | 7 | 8·5 | 0·0009 | 39·4 | 7·5 | 0·1 | 7 | 14·2 | 0·0009 | 43·0 | 15·2 | 0·3 | 0·1 | |
| Sdt8·1 + | 8 | 10·1 | 0·0009 | 20·4 | 9·5 | 0·1 | 8 | 12·1 | 0·0009 | 29·7 | 7·8 | 0·2 | 0·1 | |
| Sdt9·1 + | 9 | 2·6 | 0·0009 | 51·4 | 2·1 | 0·1 | 9 | |||||||
| Sdt10·1 + | 10 | 14·0 | 0·0009 | 38·6 | 13·8 | 0·2 | 10 | |||||||
| Sdt11·1– | 11 | 4·9 | 0·0009 | 29·2 | 4·2 | –0·1 | 11 | |||||||
| PDL | Pdl1·1 + | 1 | 21·9 | 0·0009 | 81·3 | 12·1 | 5·4 | 1 | 22·3 | 0·0009 | 86·5 | 14·3 | 3·1 | –1·2 |
| Pdl2·1 + | 2 | 2·2 | 0·0112 | 45·6 | 0·9 | 1·5 | 2 | |||||||
| Pdl3·1 + | 3 | 13·0 | 0·0009 | 1·8 | 6·6 | 4·0 | 3 | 22·8 | 0·0009 | 15·1 | 19·9 | 3·6 | 1·8 | |
| Pdl4·1 + | 4 | 6·2 | 0·0009 | 33·2 | 2·9 | 2·7 | 4 | 2·4 | 0·0280 | 38·7 | 0·9 | 0·7 | 0·7 | |
| Pdl5·1 + | 5 | 9·6 | 0·0009 | 64·9 | 4·5 | 3·3 | 5 | 7·0 | 0·0009 | 49·9 | 3·1 | 1·5 | 0·0 | |
| Pdl7·1 + | 7 | 42·4 | 0·0009 | 26·5 | 31·0 | 8·7 | 7 | 31·6 | 0·0009 | 33·9 | 26·9 | 4·4 | 0·1 | |
| Pdl8·1 + | 8 | 15·7 | 0·0009 | 20·4 | 8·5 | 4·6 | 8 | 12·3 | 0·0009 | 23·6 | 6·9 | 2·1 | 0·9 | |
| Pdl9·1– | 9 | 3·0 | 0·0019 | 29·6 | 1·3 | –1·8 | 9 | |||||||
| Pdl11·1 + | 11 | 23·0 | 0·0009 | 39·0 | 14·4 | 5·9 | 11 | 22·6 | 0·0009 | 24·4 | 10·4 | 1·7 | –3·1 | |
| PDW | Pdw1·1 + | 1 | 17·8 | 0·0009 | 111·0 | 12·1 | 0·1 | 1 | 14·7 | 0·0009 | 105·8 | 10·7 | 0·0 | 0·0 |
| Pdw2·1 + | 2 | 2 | 5·5 | 0·0009 | 39·6 | 3·1 | 0·0 | 0·0 | ||||||
| Pdw3·1 + | 3 | 16·7 | 0·0009 | 1·9 | 10·6 | 0·1 | 3 | 29·7 | 0·0009 | 14·2 | 31·1 | 0·1 | 0·0 | |
| Pdw4·1 + | 4 | 11·0 | 0·0009 | 23·5 | 6·4 | 0·1 | 4 | 12·1 | 0·0009 | 24·2 | 7·3 | 0·0 | 0·0 | |
| Pdw6·1 + | 6 | 1·7 | 0·0290 | 73·4 | 1·0 | 0·0 | 6 | |||||||
| Pdw7·1 + | 7 | 35·1 | 0·0009 | 23·2 | 31·2 | 0·1 | 7 | 15·5 | 0·0009 | 42·9 | 11·0 | 0·0 | 0·0 | |
| Pdw8·1 + | 8 | 20·3 | 0·0009 | 24·9 | 14·0 | 0·1 | 8 | 21·1 | 0·0009 | 24·6 | 16·4 | 0·1 | 0·0 | |
| Pdw11·1 + | 11 | 5·9 | 0·0009 | 32·1 | 3·5 | 0·0 | 11 | 3·3 | 0·0009 | 24·4 | 1·5 | 0·0 | 0·0 | |
| PDSBS | Pdsbs1·1 + | 1 | 17·3 | 0·0009 | 75·5 | 11·2 | 4·6 | 1 | 14·5 | 0·0009 | 64·0 | 8·8 | 1·2 | –1·6 |
| Pdsbs2·1 + | 2 | 2 | 9·2 | 0·0009 | 64·0 | 5·1 | 1·2 | 0·6 | ||||||
| Pdsbs3·1 + | 3 | 6·3 | 0·0009 | 3·3 | 3·5 | 2·6 | 3 | 21·4 | 0·0009 | 12·9 | 21·6 | 2·4 | 1·1 | |
| Pdsbs4·1 + | 4 | 6·2 | 0·0009 | 37·8 | 4·3 | 2·9 | 4 | 11·0 | 0·0009 | 0·5 | 5·2 | 1·1 | 0·9 | |
| Pdsbs5·1 + | 5 | 5 | 8·8 | 0·0009 | 71·7 | 4·5 | 1·2 | 0·0 | ||||||
| Pdsbs7·1 + | 7 | 30·4 | 0·0009 | 34·2 | 25·0 | 6·9 | 7 | 6·2 | 0·0009 | 9·0 | 5·1 | 1·2 | 0·2 | |
| Pdsbs8·1 + | 8 | 8·1 | 0·0009 | 9·6 | 5·4 | 3·2 | 8 | 16·2 | 0·0009 | 13·5 | 14·4 | 2·0 | –0·9 | |
| Pdsbs9·1 + | 9 | 9 | 7·7 | 0·0009 | 66·5 | 4·5 | 1·1 | –0·4 | ||||||
| Pdsbs11·1– | 11 | 31·4 | 0·0009 | 32·3 | 28·4 | –7·4 | 11 | 17·4 | 0·0009 | 18·1 | 10·8 | –1·1 | 2·0 | |
| STT | Stt1·1– | 1 | 4·0 | 0·0019 | 87·5 | 7·6 | –0·6 | 1 | ||||||
| Stt2·1 + | 2 | 1·7 | 0·0465 | 75·1 | 3·1 | 0·4 | 2 | 3·1 | 0·0102 | 86·9 | 4·3 | 0·6 | 0·1 | |
| Stt3·1 + | 3 | 2·8 | 0·0056 | 78·1 | 5·2 | 0·5 | 3 | 6·4 | 0·0009 | 83·2 | 9·9 | 0·9 | 0·2 | |
| Stt4·1 + | 4 | 4 | 4·1 | 0·0019 | 30·8 | 5·9 | 0·7 | 0·0 | ||||||
| Stt6·1 + | 6 | 1·8 | 0·0262 | 23·6 | 3·0 | 0·4 | 6 | 3·7 | 0·0028 | 56·0 | 5·8 | 0·6 | 0·5 | |
| Stt7·1 + | 7 | 7 | 5·8 | 0·0009 | 42·0 | 13·0 | 1·0 | 0·4 | ||||||
| Stt9·1– | 9 | 2·0 | 0·0112 | 19·5 | 4·6 | –0·5 | 9 | |||||||
| Stt10·1– | 10 | 2·2 | 0·0112 | 63·2 | 3·7 | –0·4 | 10 | |||||||
| Stt11·1– | 11 | 2·3 | 0·0065 | 35·5 | 4·1 | –0·4 | 11 | 4·7 | 0·0009 | 23·8 | 6·6 | 0·7 | –0·4 | |
| LFPL | Lfpl3·1 + | 3 | 2·0 | 0·0234 | 29·7 | 3·2 | 0·2 | 3 | ||||||
| Lfpl6·1 + | 6 | 2·0 | 0·0168 | 79·5 | 3·1 | 0·2 | 6 | |||||||
| Lfpl7·1 + | 7 | 10·9 | 0·0009 | 28·7 | 20·9 | 0·5 | 7 | |||||||
| Lfpl8·1 + | 8 | 1·7 | 0·0475 | 62·4 | 2·5 | 0·2 | 8 | |||||||
| Lfpl11·1 + | 11 | 4·9 | 0·0009 | 0·0 | 8·2 | 0·3 | 11 | 3·9 | 0·0009 | 21·4 | 10·0 | 0·3 | 0·1 | |
| LFPW | Lfpw1·1 + – | 1 | 3·1 | 0·0019 | 101·5 | 4·8 | 0·1 | 1 | 6·1 | 0·0009 | 6·7 | 9·7 | –0·2 | 0·0 |
| Lfpw2·1 + | 2 | 2 | 5·5 | 0·0009 | 53·8 | 7·6 | 0·1 | –0·1 | ||||||
| Lfpw3·1 + | 3 | 3·8 | 0·0009 | 26·6 | 5·9 | 0·2 | 3 | 6·0 | 0·0009 | 89·3 | 9·6 | 0·2 | 0·0 | |
| Lfpw6·1– | 6 | 2·9 | 0·0009 | 8·6 | 5·0 | –0·1 | 6 | |||||||
| Lfpw7·1 + | 7 | 7·3 | 0·0009 | 28·6 | 13·1 | 0·2 | 7 | 3·0 | 0·0112 | 0·0 | 5·4 | 0·1 | 0·0 | |
| Lfpw8·1 + | 8 | 2·6 | 0·0084 | 79·4 | 4·4 | 0·1 | 8 | |||||||
| Lfpw9·1– | 9 | 1·8 | 0·0215 | 38·9 | 2·7 | –0·1 | 9 | 3·8 | 0·0009 | 41·3 | 6·8 | –0·1 | 0·0 | |
| Lfpw11·1 + | 11 | 1·4 | 0·0393 | 5·7 | 2·3 | 0·1 | 11 | 5·6 | 0·0009 | 20·2 | 9·5 | 0·2 | 0·0 | |
| ECL | Ecl2·1– | 2 | 2·0 | 0·0206 | 25·6 | 1·9 | –0·2 | 2 | ||||||
| Ecl4·1 + | 4 | 6·1 | 0·0009 | 42·2 | 8·3 | 0·4 | 4 | |||||||
| Ecl6·1– | 6 | 6·8 | 0·0009 | 34·9 | 9·9 | –0·4 | 6 | |||||||
| Ecl7·1 + | 7 | 3·1 | 0·0009 | 41·0 | 3·0 | 0·2 | 7 | |||||||
| Ecl9·1– | 9 | 6·1 | 0·0009 | 33·2 | 6·9 | –0·3 | 9 | 2·3 | 0·0178 | 49·7 | 5·2 | –0·3 | 0·2 | |
| Ecl10·1 + | 10 | 22·9 | 0·0009 | 63·2 | 28·7 | 0·7 | 10 | 3·2 | 0·0056 | 59·8 | 7·6 | 0·2 | –0·6 | |
| STL10 | Stl101·1 + | 1 | 2·8 | 0·0056 | 104·6 | 2·0 | 8·8 | 1 | ||||||
| Stl102·1– | 2 | 7·9 | 0·0009 | 56·9 | 6·3 | –15·9 | 2 | |||||||
| Stl103·1 + | 3 | 3 | 7·5 | 0·0009 | 32·1 | 8·1 | 7·9 | –1·5 | ||||||
| Stl104·1 + | 4 | 4 | 4·7 | 0·0009 | 81·6 | 4·1 | 4·1 | –5·4 | ||||||
| Stl105·1– | 5 | 5·7 | 0·0009 | 54·7 | 4·2 | –12·9 | 5 | 6·0 | 0·0009 | 67·2 | 5·5 | –6·5 | 1·3 | |
| Stl106·1– | 6 | 9·5 | 0·0009 | 60·1 | 7·1 | –16·8 | 6 | 7·9 | 0·0009 | 94·9 | 8·6 | –7·3 | 5·2 | |
| Stl107·1 + | 7 | 14·6 | 0·0009 | 29·5 | 11·6 | 21·5 | 7 | 17·6 | 0·0009 | 36·9 | 19·7 | 10·5 | –9·4 | |
| Stl108·1– | 8 | 2·8 | 0·0009 | 68·2 | 1·9 | –8·7 | 8 | |||||||
| Stl109·1– | 9 | 4·0 | 0·0009 | 51·4 | 2·6 | –10·2 | 9 | 4·6 | 0·0009 | 64·3 | 3·8 | 5·3 | –1·7 | |
| Stl1010·1 + | 10 | 30·2 | 0·0009 | 59·5 | 33·9 | 36·7 | 10 | 16·1 | 0·0009 | 59·8 | 16·0 | 10·5 | –5·3 | |
| Stl1011·1 + | 11 | 4·8 | 0·0009 | 5·7 | 3·4 | 11·6 | 11 | |||||||
| STLW | Stlw1·1 + | 1 | 4·2 | 0·0009 | 62·9 | 4·6 | 19·3 | NI | ||||||
| Stlw2·1– | 2 | 5·4 | 0·0009 | 51·0 | 6·4 | –22·7 | ||||||||
| Stlw3·1– | 3 | 2·1 | 0·0158 | 3·3 | 2·3 | –13·6 | ||||||||
| Stlw5·1– | 5 | 2·7 | 0·0056 | 35·7 | 2·9 | –15·3 | ||||||||
| Stlw6·1– | 6 | 6·0 | 0·0009 | 61·2 | 6·7 | –23·3 | ||||||||
| Stlw8·1– | 8 | 4·2 | 0·0009 | 39·0 | 4·6 | –19·3 | ||||||||
| Stlw10·1 + | 10 | 19·0 | 0·0009 | 59·9 | 28·6 | 48·1 | ||||||||
| BRN | ND | NI | ||||||||||||
| FLD | Fld1·1 + | 1 | 1 | 3·7 | 0·0009 | 17·0 | 4·4 | 1·2 | –0·8 | |||||
| Fld2·1 + | 2 | 2 | 15·7 | 0·0009 | 65·5 | 17·6 | 2·5 | –0·6 | ||||||
| Fld4·1 + – | 4 | 1·9 | 0·0177 | 3·1 | 2·6 | 1·4 | 4 | 3·7 | 0·0056 | 81·6 | 2·9 | –1·0 | –0·4 | |
| Fld5·1– | 5 | 5 | 7·8 | 0·0009 | 50·3 | 6·4 | –0·6 | 2·0 | ||||||
| Fld6·1 + | 6 | 6 | 4·8 | 0·0009 | 36·6 | 6·1 | 0·7 | –1·9 | ||||||
| Fld7·1 + | 7 | 5·3 | 0·0009 | 54·1 | 7·7 | 2·4 | 7 | 5·1 | 0·0009 | 98·7 | 6·9 | 1·6 | –0·3 | |
| Fld8·1 + | 8 | 5·3 | 0·0009 | 36·1 | 7·9 | 2·4 | 8 | 5·1 | 0·0009 | 55·1 | 4·8 | 1·3 | –0·4 | |
| Fld9·1 + | 9 | 2·7 | 0·0047 | 50·8 | 3·6 | 1·6 | 9 | 2·9 | 0·0065 | 0·0 | 2·3 | 0·6 | 1·0 | |
| Fld10·1 + | 10 | 3·0 | 0·0056 | 47·8 | 4·0 | 1·7 | 10 | 13·1 | 0·0009 | 39·2 | 15·6 | 1·7 | –2·4 | |
| Fld11·1– | 11 | 11·4 | 0·0009 | 40·4 | 19·6 | –3·8 | 11 | |||||||
| PDDM | Pddm1·1– | 1 | 1 | 5·5 | 0·0009 | 28·3 | 9·2 | –1·0 | –0·4 | |||||
| Pddm2·1 + | 2 | 2 | 2·7 | 0·0206 | 78·3 | 5·5 | 0·8 | –0·2 | ||||||
| Pddm3·1 + | 3 | 3 | 4·4 | 0·0009 | 2·0 | 17·1 | 1·4 | –0·1 | ||||||
| Pddm4·1 + | 4 | 2·7 | 0·0009 | 52·9 | 6·6 | 0·9 | 4 | |||||||
| Pddm6·1 + | 6 | 2·5 | 0·0037 | 57·7 | 5·1 | 0·8 | 6 | |||||||
| Pddm7·1 + | 7 | 3·8 | 0·0009 | 59·7 | 7·9 | 1·0 | 7 | 2·8 | 0·0178 | 13·5 | 5·5 | 0·3 | –1·0 | |
| SDTWT | Sdtwt2·1 + | 2 | ND | 2 | 3·2 | 0·0019 | 37·1 | 4·5 | 4·6 | 43·5 | ||||
| Sdtwt4·1 + | 4 | 4 | 2·7 | 0·0122 | 50·0 | 4·7 | 28·4 | 20·0 | ||||||
| Sdtwt5·1 + | 5 | 5 | 2·8 | 0·0168 | 49·9 | 3·6 | 27·0 | –10·2 | ||||||
| Sdtwt6·1 + | 6 | 6 | 3·9 | 0·0028 | 52·3 | 5·1 | 9·4 | 45·0 | ||||||
| Sdtwt7·1 + | 7 | 7 | 8·0 | 0·0009 | 48·5 | 23·1 | 64·5 | 39·9 | ||||||
| Sdtwt10·1– | 10 | 10 | 4·1 | 0·0009 | 38·6 | 6·6 | –22·5 | 42·9 | ||||||
| SDNPPD | Sdnppd7·1 + | 7 | 7 | 10·6 | 0·0009 | 45·9 | 12·8 | 1·5 | –0·4 | |||||
| Sdnppd11·1 + | 11 | 48·1 | 0·0009 | 36·5 | 70·1 | 5·4 | 11 | 31·6 | 0·0009 | 26·6 | 45·7 | 1·7 | –3·2 | |
| PDTN | Pdtn1·1– | 1 | 3·9 | 0·0009 | 91·6 | 5·7 | –3·9 | 1 | ||||||
| Pdtn2·1– | 2 | 2 | 4·0 | 0·0028 | 73·7 | 1·8 | –50·3 | 47·6 | ||||||
| Pdtn3·1– | 3 | 3·3 | 0·0019 | 0·0 | 4·7 | –3·5 | 3 | |||||||
| Pdtn4·1– | 4 | 2·4 | 0·0047 | 64·0 | 3·6 | –3·1 | 4 | |||||||
| Pdtn6·1 + | 6 | 6 | 11·0 | 0·0009 | 10·6 | 57·1 | 270·1 | –283·0 | ||||||
| Pdtn7·1– + | 7 | 3·5 | 0·0009 | 28·0 | 5·2 | –3·7 | 7 | 7·1 | 0·0009 | 50·2 | 8·1 | 69·6 | 149·3 | |
| Pdtn8·1– | 8 | 2·2 | 0·0178 | 70·1 | 3·2 | –2·9 | 8 | 10·3 | 0·0009 | 10·2 | 6·7 | –107·2 | 59·8 | |
| Pdtn9·1– | 9 | 1·8 | 0·0122 | 6·9 | 3·5 | –3·0 | 9 | |||||||
| Pdtn10·1– | 10 | 10 | 3·9 | 0·0009 | 38·6 | 2·0 | –57·9 | 33·9 | ||||||
| Pdtn11·1– | 11 | 9·4 | 0·0009 | 35·8 | 15·6 | –6·4 | 11 | 12·5 | 0·0009 | 3·5 | 6·8 | –114·6 | 26·1 | |
NI, not investigated; ND, no significant QTL detected.
Seed coat permeability (SDP)
One key trait in domestication is reduction or loss of seed dormancy, which enables uniform germination. Like several cereal and legume crops, domestication of cowpea/yardlong bean has resulted in reduced seed dormancy. SDP in yardlong bean and wild cowpea were 100 and 0 %, respectively. Six QTLs were detected on LGs 1, 2, 4, 7, 8 and 11, where all alleles, except Sdp7·1–, of the yardlong bean parent increase the percentage of permeable seeds. The QTL located on LG1 had the largest effect on phenotypic variance explained (Table 5).
Pod dehiscence (PDT and PDD)
Loss of pod dehiscence in legume crops is advantageous for harvesting seeds but reducing seed dispersal in wild species. Pod dehiscence can be characterized as a qualitative or quantitative trait based on visual score (PDD; dehiscent vs. indehiscent) or number of twists along the shattered pod (PDT).
The qualitative nature of dehiscence was examined only in the F2 population (Table 3) and the progenies segregated for PDD in the ratio 111 dehiscent to 77 indehiscent plants. The segregation ratio was 9 : 7 (χ2 = 0·60, P = 0·44), indicating that duplicated recessive genes control pod indehiscence.
The quantitative nature of pod dehiscence was investigated only in the BC1F1 population (Table 4). Yardlong bean JP81610 showed no pod twisting in contrast to wild cowpea TVnu457. Four QTLs were detected on LGs 1, 4, 7 and 9. Of these QTLs, the one on LG7 had the largest effect on phenotypic variance. Alleles of the cultivated parent reduced the number of twists on the pod at all QTLs.
Organ size (seed size, pod size, spacing between seeds and stem thickness)
Domestication of yardlong bean resulted in an approx. eight-fold increase in seed weight (Table 3). Eight to ten QTLs for traits related to seed size (SD100WT, SDL, SDW and SDT) were located on all LGs. At all QTLs, alleles from the cultivated parent increases the size of each trait, except alleles of the QTLs on LG11, which decreased SD100WT, SDL and SDT. The QTLs with the largest phenotypic contribution for SD100WT (24·6 %), SDL (26·5 %) and SDW (20·9 %) were located on LG7, and that for SDT (26·4 %) was located on LG3 (Table 5).
The most remarkable domesticated trait of yardlong bean is its pod length. Domestication of yardlong bean has resulted in an approx. eight-fold increase in pod length. Seven to nine QTLs were identified for pod-related traits on LGs 1, 2, 3, 4, 5, 7, 8, 9 and 11 (Table 5). The alleles from yardlong bean increased pod size at almost all QTLs. Alleles from wild cowpea reduced the value of pod size only on LG9 for PDL and on LG11 for PDSBS. The QTLs located on LG7 had the greatest effect for PDL and PDW. For PDSBS, the QTLs showing the largest effect were located on LGs 3, 7 and 11. Generally, QTLs for pod size were clustered with or located close to QTLs for seed size-related traits, especially the QTLs on LGs 3, 4, 7, 8 and 11 (Fig. 2). The QTLs on LG7 for PDL, PDW and PDSBS were also located near to the QTL for pod dehiscence.
Fig. 2.
QTLs detected on BC1F1 (upper) and F2 (lower) populations from the cross between cultivated yardlong bean and wild cowpea. The effect of the cultivated parent is indicated after each QTL name. For explanation of trait abbreviations, see Table 1.
Four QTLs for leaf length were found on LGs 3, 6, 7 and 11 in the BC1F1 population. However, only that on LG11 was confirmed in the F2 population. For leaf width, nine QTLs were detected on LGs 1, 2, 3, 6, 7, 8, 9 and 11. In both traits, the QTLs on LG7 had the greatest effect and were at the same position (Table 5, Fig. 2).
Nine QTLs for stem thickness were detected on LGs 1, 2, 3, 4, 6, 7, 9, 10 and 11. The QTL on LG7 had the greatest effect. Except for the QTLs on LGs 1, 9, 10 and 11, the cultivated alleles at these QTLs increased STT.
Growth habit (ECL, STL10 and STLW)
Epicotyl length, stem length within the first ten internodes and whole stem length (ECL, STL10 and STLW) in the cultivated parents were higher than those in the wild parent. Six QTLs for ECL were found on LGs 2, 4, 6, 7, 9 and 10. The cultivated alleles at QTLs Ecl2·1-, Ecl6·1- and Ecl9.1- decreased epicotyl length. Eleven QTLs for STL10 were detected on all LGs. The cultivated alleles at QTLs Stl102·1–, Stl105·1–, Stl106·1–, Stl108·1– and Stl109·1– lowered internode length. Seven QTLs for STLW were identified on LGs 1, 2, 3, 5, 6, 8 and 10. At QTLs Stlw2·1–, Stl3·1–, Stlw5·1–, Stlw6·1– and Stlw8·1, the alleles from the cultivated parent decreased upper internode length. For all growth habit traits, the QTLs on LG10 had the greatest effect, with alleles increasing trait values. The QTLs for epicotyl and internode lengths were located on the same or similar position on LGs 5, 7 and 10. QTLs for lower and upper internode lengths were also mapped to the same or similar location on LGs 2, 6 and 10.
Flowering time (FLD)
Days to first flowering of the cultivated and wild parents were almost the same (58 vs. 57 d) in Japan, but relatively different (55 vs. 44 d) in Thailand. However, large variation existed in the two populations. Ten QTLs were detected on all LGs except LG3 for FLD. QTLs on LGs 2, 10 and 11 had a large effect on phenotypic variance. Unexpectedly, alleles from the cultivated parent at all QTLs except LG5 and LG11 delayed flowering.
Maturity time (PDDM)
The cultivated parent had a much larger seed and pod than the wild parent. Yardlong bean would be expected to take longer to reach pod maturity than the wild parent due to greater translocation of dry matter to seeds and pods. Six QTLs associated with days to maturity were found on LGs 1, 2, 3, 4, 6 and 7. The alleles from the cultivated parent that delayed maturity were on LGs 2, 3, 4, 6 and 7, whereas a QTL on LG1 hastened maturity. The QTL on LG3 had the greatest effect and was located near the QTLs for seed- and pod-related traits.
Yield-related traits (PDTN, SDNPPD and SDTWT)
The cultivated parent produced a markedly lower number of pods than the wild parent. Ten QTLs were detected on all LGs, except LG5. Of these QTLs, only at LG6 did the allele from yardlong bean increase PDTN. This QTL had the largest effect, accounting for 57·1 % of the phenotypic variation.
Although yardlong bean has much longer pods than wild cowpea, the average seed number per pod was only slightly different (three seeds). Two QTLs were found on LGs 7 and 11 for SDNPPD. The QTL on LG11 had largest effect, explaining 70 % of the phenotypic variation in the BC1F1 population. This QTL was co-located with QTLs for flowering time, stem thickness, leaf size, and seed- and pod-related traits. As expected, the alleles from the yardlong bean parent increased the number of seeds per pod at both QTLs (Table 5, Fig. 2).
The wild parent has smaller seed size but higher total seed weight than the cultivated parent. SDTWT was investigated only in the F2 population. Six QTLs were found on LGs 2, 4, 5, 6, 7 and 10. The QTL with the largest effect was located on LG7. This QTL was clustered with stem-, seed- and pod-related traits. The alleles from the cultivated parent at all QTLs increased total seed weight.
Seed coat colour (SDC)
Seed coat colour was characterized as a qualitative trait. The cultivated parent had a black seed coat whereas the wild parent had a brown seed coat. F2 progeny segregated for seed coat colour at a ratio of 33 (brown) to 155 (black). Although the segregation ratio fitted to 3 : 13 (χ2 = 0·18, P = 0·67), it also fitted to 1 : 3 at P = 0·01 (χ2 = 5·56, P = 0·018). The BC1F1 progenies showed no segregation and all had a black seed coat. These suggested that seed coat colour may be governed by dominant and recessive epistasis or by a single gene and that black is dominant over brown. However, when this trait was mapped as a morphological marker, seed coat colour was located near to marker cp03855, which showed high segregation distortion.
Distribution of domestication trait-related QTLs
Although it is not known whether all the 153 QTLs detected for domestication-related traits had independent actions on each trait, the observed number of QTLs was compared with the expected number based on each LG length (Table 6). The observed and expected numbers agreed well throughout all LGs, except for LG11 for which the number of QTLs was statistically higher than expected. The departure in LG11 caused significant differences in total χ2 values for both F2 and BC1F1 at 17·5 and 17·8, respectively.
Table 6.
Observed and expected numbers of QTLs and their chi-square values on each linkage groups
|
BC1F1 |
F2 |
|||||||
|---|---|---|---|---|---|---|---|---|
| LG | Length (cM) | Detected QTLs | Expected QTLs | χ2 | Length (cM) | Detected QTLs | Expected QTLs | χ2 |
| LG1 | 147·3 | 14 | 20·9 | 2·3 | 140·4 | 11 | 15·2 | 1·2 |
| LG2 | 76·4 | 8 | 10·8 | 0·7 | 93·4 | 12 | 10·1 | 0·3 |
| LG3 | 78·8 | 10 | 11·2 | 0·1 | 105·7 | 11 | 11·5 | 0·0 |
| LG4 | 67·4 | 14 | 9·6 | 2·0 | 87·2 | 11 | 9·5 | 0·3 |
| LG5 | 64·9 | 7 | 9·2 | 0·5 | 73·7 | 6 | 8·0 | 0·5 |
| LG6 | 79·5 | 10 | 11·3 | 0·1 | 107·7 | 7 | 11·7 | 1·9 |
| LG7 | 85·6 | 15 | 12·1 | 0·7 | 107·1 | 16 | 11·6 | 1·7 |
| LG8 | 86·0 | 14 | 12·2 | 0·3 | 103·4 | 10 | 11·2 | 0·1 |
| LG9 | 60·3 | 8 | 8·6 | 0·0 | 70·5 | 6 | 7·6 | 0·4 |
| LG10 | 63·2 | 7 | 9·0 | 0·4 | 59·8 | 7 | 6·5 | 0·0 |
| LG11 | 43·1 | 14 | 6·1 | 10·2** | 28·3 | 9 | 3·1 | 11·5*** |
| Total | 852·4 | 121 | 121 | 17·5*** | 977·1 | 106 | 106 | 17·8*** |
Asterisks indicate significant differences between the observed and expected numbers of QTLs in each linkage group at **P < 0·01 and ***P < 0·001.
DISCUSSION
Linkage maps of yardlong bean
The yardlong bean QTL genetic maps presented here are the first for comprehensive domestication-related traits derived from an inter-subspecific cross of this crop. The number of linkage groups corresponded to the haploid chromosome number (n = 11) of yardlong bean (cowpea). In this study, 113 SSR markers derived from cowpea, azuki bean and mungbean were chosen from our previous map constructed from a BC1F1 population (Kongjaimun et al., 2012), and were integrated into the F2 genetic map. The LG and marker orders on each LG have been confirmed. Therefore, these yardlong bean maps are useful for understanding genome synteny among Vigna species, as well as for identifying QTLs for useful traits.
Some genetic barriers exist in inter-subspecific crosses between cowpea and its wild progenitor. Ng (1995) reported partial incompatibility between cultivated cowpea V. unguiculata ssp. unguiculata and wild cowpea V. unguiculata ssp. dekindtiana. In our study, in which yardlong bean and wild cowpea were crossed, 27 plants in the F2 population did not set pod, and 15 had a very low pod set (≤15) or had shrivelled seeds. In the BC1F1 population, one plant failed to produce any pods and nine plants produced a small number of pods (ten or fewer). Those plants were excluded from the mapping population.
Markers showing segregation distortion were observed on some LGs on both the F2 and the BC1F1 maps. The F2 map had more distorted markers than the BC1F1 map (P < 0·05, 48·7 vs. 9·7 %). In particular, the markers located on LG11, where 7/9 and 13/14 of the F2 and BC1F1 maps, respectively, showed distorted segregation. The direction of skewed markers on LG11 in both maps was toward the homozygous yardlong bean genotype. The same results have been found in an F2 interspecific linkage map of V. umbellata × V. nakashimae (Somta et al., 2006), and in a BC1F1 linkage map of V. umbellata (Isemura et al., 2010). They reported that none out of ten markers and all markers on LG11 of the respective linkage maps were highly distorted. The cluster of distorted markers may result from the presence of a gene(s) controlling sterility and/or compatibility (Zamir and Tadmor, 1986). Therefore, a gene(s) on LG11 may play an important role in genetic differentiation, both within yardlong bean (cowpea) and among Vigna species.
Highly distorted segregation may affect inheritance of nearby traits, even with simply inherited traits. In mungbean, Lambrides et al. (2004) found that the segregation ratio suggested dominant and recessive epistasis controlling seed coat colour. However, this was rejected and a single-gene model was accepted based on molecular marker data. They eventually mapped seed coat colour locus to a distorted genomic region with an understanding that the aberrant segregation ratio was due to the effect of segregation distortion. In our study on seed coat colour, segregation ratios suggest either dominant and recessive epistasis or single gene control, although gene mapping supported the latter.
The genetics of domestication-related traits
In yardlong bean the majority of domestication-related traits are controlled by one major QTL and several minor QTLs, and a few are controlled by single major genes. Results from this study revealed that yardlong bean differs from other Vigna crops in the genetic control of some simply inherited traits. For example, pod dehiscence, which is controlled by a single gene in azuki bean (Isemura et al., 2007; Kaga et al., 2008) and rice bean (Isemura et al. 2010), is controlled by several genes/QTLs in yardlong bean. Nonetheless, in a related crop, common bean (Phaseolus vulgaris) pod dehiscence is also governed by several genes (Koinange et al., 1996).
In most of the domestication-related traits measured, 2–11 QTLs on two or more linkage groups were detected. QTLs for many related traits such as pod size, seed size, leaf size and stem-related traits were co-located on the same position on the same linkage groups. The same findings were reported for azuki bean (Isemura et al., 2007; Kaga et al., 2008) and rice bean (Isemura et al., 2010). Co-location of QTLs for these traits may be due to the effect of developmental allometry (Smartt, 1976).
Genomic regions and distribution of QTLs for domestication-related traits
The QTLs controlling domestication traits are generally not randomly distributed across the crop genome (Gepts, 2004). The non-randomness of the domestication QTLs may be related to ‘cultivation magnetism’ and should be considered under the ‘protracted transition paradigm’ of crop domestication (Allaby, 2010). Domestication-related traits have been studied in several legume crops such as common bean (Koinange et al., 1996), azuki bean (Isemura et al., 2007; Kaga et al., 2008), rice bean (Isemura et al., 2010) and soybean (Liu et al., 2007); the results revealed that domestication traits are controlled by several QTLs/genes with each trait controlled by a few large QTLs or by a major QTL and several minor QTLs. The results from our study demonstrated that for most traits the latter case exists in yardlong bean. For example, one QTL with large effect and 3–9 minor QTLs control SDP, SDT, SD100WT, PDL, PDW, STL10 and STLW.
The distribution of domestication-related QTLs across the yardlong bean genome is shown by co-location of the QTLs on several narrow genomic regions on almost all linkage groups, especially LG3, LG7, LG8 and LG11 (Fig. 2). LG3 is associated with QTLs for increased organ size, such as seed, pod and leaf, which were detected in a limited region of 16 cM. LG7 is associated with pod dehiscence, organ size (seed, pod, stem and leaf), stem length and yield potential. Major QTLs for organ size were principally located on this LG. In addition, the QTLs for days to first flower and days to maturity of first pod are closely linked in a region of 6 cM on LG7. LG8 is associated with increased seed and pod size. LG11 is associated with organ size (seed, pod, stem and leaf), earliness and yield potential traits.
QTL clusters for domestication are reported in several crops. Clustering of QTLs is due to either close linkage or pleiotropy or both. Single mutations can have pleiotropic effects on various organs. In common bean, the determinacy gene (fin) has pleiotropic effects on the number of nodes on the main stem, the number of pods, and the number of days to flowering and maturity (Koinange et al., 1996). In maize, QTLs related to change in inflorescence sex, and number and length of internodes in lateral branches and inflorescences are distributed within a narrow genomic region (Doebley et al., 1995). Change in these traits is explained by the pleiotropic effect of a single tb1 gene. In tomato, the seed testa colour mutant gene bks decreases seed weight and increases fruit pH (Downie et al., 2003). In sunflower, the unbranched allele (B) decreased seed oil content and increased capitula diameter and seed weight (Bachlava et al., 2010).
Comparison of domestication QTLs between yardlong bean and related Vigna and other crops
QTLs with larger effect [≥20 % phenotypic variation explained (PVE)] are considered here.
Comparison with azuki bean
Distribution of the main QTLs between the two species showed a marked difference. In yardlong bean, large-effect QTLs were detected on seven linkage groups – LGs 1, 3, 6, 7, 8, 10 and 11. In azuki bean, large-effect QTLs were found on only five linkage groups – LGs 1, 2, 7, 8 and 9 (Isemura et al., 2007; Kaga et al., 2008). Marked differences between yardlong bean and azuki bean were found on LG6 and LG7. Several large-effect QTLs were detected on LG6 of azuki bean, whereas only one QTL was detected on this LG in yardlong bean. Major QTLs for domestication-related traits were abundant on LG7 in yardlong bean, but only a few QTLs were detected in azuki bean (Supplementary Data, Table S4).
Seed size. Seed weight of the mapping parents used in this study showed an eight-fold variance. The 100-seed weight of cultivated yardlong bean was 19·4 g while that of the wild parent was 2·5 g. Ten QTLs were detected for 100-seed weight and that with the largest effect (PVE = 21·1 %) was found on LG7 for yardlong bean, whereas eight QTLs for this trait were detected in azuki bean that with the largest effect (PVE = 31·3 %) was located on LG2. QTLs for 100-seed weight in yardlong bean and azuki bean on LG1, LG10 and LG11 may be common. QTLs for seed size in both crops on LG1 were linked to markers CEDG090 and CEDG051, on LG10 were between markers CEDG113 and CEDG106, and on LG11 were linked to marker CEDG076.
Pod dehiscence. Reducing seed dispersal is one of the first steps in crop domestication (Ladizinsky, 1979). Pod dehiscence results from the presence of fibres surrounding the vascular bundles in the pod walls and a fibrous parchment layer lining the pod cavity (Roth, 1977). Pod twisting is caused by the oblique orientation of the fibres in the parchment layer. Four QTLs were detected for pod dehiscence in yardlong bean. The QTL with the highest contribution (48 %) was found on LG7 whereas a single QTL for this trait with a higher contribution (90·5 %) was detected on the same LG7 in azuki bean (Isemura et al., 2007). The pod size QTL with the largest effect in azuki bean and yardlong bean were both found on LG7.
Pod length. Among the pod-related traits, length of pod is the main distinguishing trait of yardlong bean. The distinct pod character of yardlong bean may be because it has been domesticated solely for edible young pods. The domesticated yardlong bean parent used here had a pod length of 78·1 cm, while domesticated azuki bean had a pod length of 9·2 cm (Isemura et al., 2007). The yardlong bean and wild cowpea parents in this study showed an eight-fold difference in pod length, whereas azuki bean and its wild relative have similar pod length (Isemura et al., 2007). Nine QTLs were involved in pod length in yardlong bean. Five were associated with the same trait in azuki bean. Largest-effect QTLs for the two species were detected on LG7. These QTL and that on LG1 were considered to be common in both Vigna crops.
Seed dormancy. Physical seed dormancy is generally caused by the presence of water-impermeable layers of palisade cells in the seed coat (Finch-Savage and Leubner-Metzger, 2006). Water absorption occurs through the structure of the strophiole (parenchymatous tissue) adjacent to the hilum in Vigna species (Gopinathan and Babu, 1985). Six QTLs were identified for seed dormancy-related traits in yardlong bean whereas two to five were identified in azuki bean. The largest-effect QTLs for dormancy-related traits in yardlong bean and azuki bean were both found on LG1 and were likely to be common as they are linked to the same markers, CEDG256 and CEDG214.
Comparison with rice bean
In rice bean, large-effect QTLs were found on three linkage groups (Isemura et al., 2010): LG2 and LG4 for seed and pod size, LG4 for water absorption by seeds (seed dormancy), and LG7 for pod dehiscence and growth habit (stem length). Differences between yardlong bean and rice bean were found on LG4 and LG7. Several QTLs with large effect were detected on LG7 of yardlong bean, whereas a few QTLs were detected on this LG7 of rice bean. In contrast, major QTLs for domestication-related traits were found on LG4 of rice bean while no major QTL was detected on LG4 of yardlong bean (Supplementary Data, Table S4).
Seed size. A QTL for 100-seed weight on LG1 of rice bean and azuki bean was common (Isemura et al., 2010), and thus this QTL of rice bean may be the same as that for seed weight in yardlong bean (see discussion above regarding yardlong bean and azuki bean).
Pod dehiscence. A major difference seen between yardlong bean and rice bean is pod dehiscence. Only a single major QTL (42·4 % PVE) controlling pod dehiscence was found on LG7 in rice bean, but one major and six minor QTLs were detected for this trait in yardlong bean. The major QTLs in both rice bean and yardlong bean were located on LG7, and are possibly the same. Marker CEDG111 is linked to both QTLs on LG7 of rice bean and yardlong bean (18·5 and 17·2 cM, respectively).
Pod length. As in azuki bean, pod length of rice bean and its wild relative was only slightly different (Isemura et al., 2010). Nine and three QTLs were detected for pod length in yardlong bean and rice bean, respectively. QTLs on LG2 and LG4 were considered common in both species. The two QTLs were linked to markers cp08299 and CEDG062, respectively. However, the largest-effect QTL in yardlong bean was on LG7 while that of rice bean was on LG4.
Seed dormancy. Six seed dormancy-related QTLs were detected in yardlong bean. Of these, five were detected in rice bean. Two QTLs for yardlong bean each on LG4 and LG8 appear to be common to rice bean. The QTL mapped on LG4 of yardlong bean and rice bean were both linked to marker CEDG062 (22 and 19·7 cM, respectively). The QTL on LG8 of the two species was linked to marker cp10549 at 1·2 and 6·1 cM, respectively.
Comparison with cowpea and mungbean: seed size
Seed weight is an important trait related to yield in domesticated Vigna. QTLs for seed weight of cowpea and mungbean have been reported by Fatokun et al. (1992), two QTLs for cowpea and four for mungbean. LGs ii and vi in the cowpea map correspond to LG1 and LG4, respectively, in the azuki bean map (Isemura et al., 2007), which also correspond to the same linkage groups in the yardlong bean map. LGs i, ii, iii and vi in the mungbean map correspond to LGs 9, 1, 8 and 10, respectively, in the azuki bean map (Isemura et al., 2007) and yardlong bean map in our study.
In this study, a QTL for seed weight was detected on LG1 at the location corresponding to that of a QTL for this trait on LG ii in cowpea and mungbean. Other QTLs for seed weight were also detected at similar locations on LG4 of yardlong bean and LG vi of cowpea, as well as LG8 and LG10 of yardlong bean and LG iii and vi of mungbean. Although the QTL with the largest effect for seed weight was detected on LG7 in yardlong bean, no QTL was detected on the corresponding linkage groups in cowpea and mungbean.
Conclusions
This is the first report of QTLs for domestication-related traits in yardlong bean/cowpea. Twenty-three domestication-related traits (three qualitative and 21 quantitative traits) were investigated in the F2 and BC1F1 populations. The 21 quantitative traits were dissected into 153 QTLs. The differences between wild cowpea and cultivated yardlong bean were found to be controlled by several major QTLs. Major QTLs for unrelated organs were distributed in clusters on LGs 3, 7 and 11. This study revealed that domestication-related traits are controlled by a few major and many minor QTLs. Comparing these results with other domesticated Vigna species showed similarity to azuki bean but differences from rice bean. High genome synteny in the genus Vigna enabled QTL comparison between yardlong bean and azuki bean, rice bean, mungbean and cowpea. Major QTLs in yardlong bean were found on LG7, whereas those of azuki bean were on LG9 and for rice bean were on LG4. Some genomic regions for seed dormancy, pod dehiscence and seed and pod size are conserved between yardlong bean and azuki bean and/or rice bean. Some genomic regions for seed size are conserved between yardlong bean, cowpea and/or mungbean. The results provide a foundation for marker-assisted selection of domestication-related QTLs in yardlong bean and enhance our understanding of domestication in the Vigna.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This study was financially supported by the Royal Golden Jubilee PhD programme (RGJ) of the Thailand Research Fund (TRF).
LITERATURE CITED
- Allaby R. Integrating the processes in the evolutionary system of domestication. Journal of Experimental Botany. 2010;61:935–944. doi: 10.1093/jxb/erp382. [DOI] [PubMed] [Google Scholar]
- Andargie M, Pasquet RS, Gowda BS, Muluvi GM, Timko MP. Construction of a SSR-based genetic map and identification of QTL of domestication traits using recombinant inbred lines from a cross between wild and cultivated cowpea (V. unguiculata (L.) Walp) Molecular Breeding. 2011;28:413–420. [Google Scholar]
- Bachlava E, Tang S, Pizarro G, et al. Pleiotropy of the branching locus (B) masks linked and unlinked quantitative trait loci affecting seed traits in sunflower. Theoretical and Applied Genetics. 2010;120:829–842. doi: 10.1007/s00122-009-1212-1. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C. Teosinte branched and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie AB, Zhang D, Dirk LMA, et al. Communication between the maternal testa and the embryo and/or endosperm affect testa attributes in tomato. Plant Physiology. 2003;133:145–160. doi: 10.1104/pp.103.022632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatokun CA, Menancio-Hautea DI, Danesh D, Young ND. Evidence for orthologous seed weight genes in cowpea and mungbean based on RFLP mapping. Genetics. 1992;132:841–846. doi: 10.1093/genetics/132.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytologist. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Fuller D, Harvey EL. The archaeobotany of Indian pulses: identification, processing and evidence for cultivation. Environmental Archaeology. 2006;11:219–246. [Google Scholar]
- Gepts P. Crop domestication as a long time selection experiment. Plant Breeding Reviews. 2004;24:1–44. [Google Scholar]
- Gopinathan MC, Babu CR. A unique growth pattern associated with cleistogamy in a tropical legume, Vigna minima (Roxb.) Ohwi & Ohashi (Leguminosae) Botanical Journal of the Linnean Society. 1985;92:263–268. [Google Scholar]
- Hammer K. Das Domestikationssyndrom. Genetic Resources and Crop Evolution. 1984;32:11–34. [Google Scholar]
- Izawa T, Konishi S, Shomura A, Yano M. DNA changes tell us about rice domestication. Current Opinion in Plant Biology. 2009;12:185–192. doi: 10.1016/j.pbi.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Isemura T, Kaga A, Konishi S, et al. Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and their comparison with other warm season legumes. Annals of Botany. 2007;100:1053–1071. doi: 10.1093/aob/mcm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemura T, Kaga A, Tomooka N, Shimizu T, Vaughan DA. The genetics of domestication of rice bean, Vigna umbellata. Annals of Botany. 2010;106:927–944. doi: 10.1093/aob/mcq188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaga A, Isemura T, Tomooka N, Vaughan DA. The genetics of domestication of the azuki bean (Vigna angularis) Genetics. 2008;178:1013–1036. doi: 10.1534/genetics.107.078451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinange EMK, Singh SP, Gepts P. Genetic control of the domestication syndrome in common bean. Crop Science. 1996;36:1037–1045. [Google Scholar]
- Kongjaimun A, Kaga A, Tomooka N, et al. An SSR-based linkage map of yardlong bean (Vigna unguiculata (L.) Walp. subsp. unguiculata Sesquipedalis Group) and QTL analysis of pod length. Genome. 2012;55:81–92. doi: 10.1139/g11-078. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Annals of Eugenics. 1944;12:172–175. [Google Scholar]
- Ladizinsky G. Seed dispersal in relation to the domestication of Middle East legumes. Economic Botany. 1979;33:284–289. [Google Scholar]
- Lambrides CJ, Godwin ID, Lawn RJ, Imrie BC. Segregation distortion for seed testa color in mungbean (Vigna radiata L. Wilczek) Heredity. 2004;95:532–535. doi: 10.1093/jhered/esh078. [DOI] [PubMed] [Google Scholar]
- Liu XB, Herbert SJ, Zhang QY, Hashemi M. Yield-density relation of glyphosate-resistant soya beans and their responses to light enrichment in north-eastern USA. Journal of Agronomy and Crop Science. 2007;193:55–62. [Google Scholar]
- Lodhi MA, Ye GN, Weeden NF, Reisch BI. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Molecular Biology Reporter. 1994;12:6–13. [Google Scholar]
- Maréchal R, Mascherpa JM, Stainer F. Etude taxonomique d'un groupe complexe d'speces des genres Phaseolus et Vigna (Papilionaceae) sur la base de donnees morphologiques et polliniques, traitees par l'analyse informatique. Boissiera. 1978;28 [Google Scholar]
- Ng NQ. Cowpea. In: Smart J, Simmonds NW, editors. Evolution of crop plants. 2nd edn. Harlow, UK: Longman Scientific and Technical; 1995. pp. 326–332. [Google Scholar]
- Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- Roth I. Fruits of angiosperms. Berlin: Borntraeger; 1977. [Google Scholar]
- Somta P, Kaga A, Tomooka N, et al. Development of an interspecific Vigna linkage map between Vigna umbellata (Thunb.) Ohwi & Ohashi and V. nakashimae (Ohwi) Ohwi & Ohashi and its use in analysis of bruchid resistance and comparative genomics. Plant Breeding. 2006;125:77–84. [Google Scholar]
- Smartt J. Tropical pulses. London: Longman; 1976. [Google Scholar]
- Smartt J. Grain legumes: evolution and genetic resources. Cambridge: Cambridge University Press; 1990. [Google Scholar]
- Tomooka N, Yoon MS, Doi K, Kaga A, Vaughan DA. AFLP analysis of diploid species in the genus Vigna subgenus Ceratotropis. Genetic Resources and Crop Evolution. 2002;49:521–530. [Google Scholar]
- Van Ooijen JW. JoinMap Version 4·0, Software for the calculation of genetic linkage maps. Wageningen: Kyazma B.V; 2006. [Google Scholar]
- Wang XW, Kaga A, Tomooka N, Vaughan DA. The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean. [Vigna angularis (Wild.) Ohwi & Ohashi]. Theoretical and Applied Genetics. 2004;109:352–360. doi: 10.1007/s00122-004-1634-8. [DOI] [PubMed] [Google Scholar]
- Xu P, Hu T, Yang Y, et al. Mapping genes governing flower and seedcoat color in asparagus bean (Vigna unguiculata ssp. sesquipedalis) based on single nucleotide polymorphism and simple sequence repeat markers. HortScience. 2011;46:1102–1104. [Google Scholar]
- Zamir D, Tadmor Y. Unequal segregation of nuclear gene in plant. Botanical Gazette. 1986;145:355–358. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.