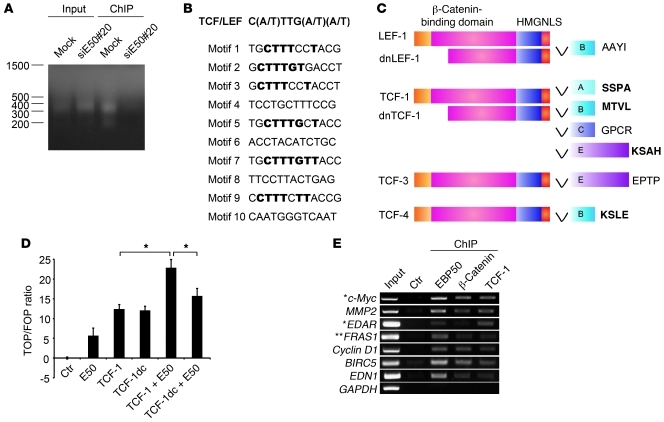
Figure 3. EBP50 binds to the TCF/LEF consensus motif.
(A) Mock- and an EBP50 siRNA–transfected SW480 clone (siE50#20) were processed for ChIP assay using anti-EBP50 antibody. The ligation-mediated PCR (LM-PCR) products from genomic (input) and anti-EBP50 antibody immunoprecipitated (ChIP) DNA are shown. (B) Ten motifs with the highest scores, compiled from the 57 most significantly EBP50-bound regions, as predicted by a BioProspector Search, are shown. Among them, 6 motifs identical or similar to the TCF/LEF consensus motif are shown in bold. (C) Molecular structure of TCF family proteins. HMG, high-mobility group proteins; NLS, nucleus localization signal. The last 4 amino acids at the C-terminal ends are shown in bold if those sequences match to a potential PDZ-binding motif. (D) SW480 cells were transfected with 2 μg of the indicated plasmids, using empty vector as a control, and 0.4 μg of Top/Fop-flash reporter plasmids. TCF-1dc is a C-terminally deleted TCF-1 construct lacking the EBP50 binding motif. Luciferase activity was assessed and presented as in Figure 2C. *P < 0.05. (E) SW480 cells were processed for ChIP assay using the indicated antibodies or a rabbit anti-podocalyxin antibody as a control. Selective Wnt/β-catenin–regulated gene sequences were amplified by PCR. *Genes reproducibly identified as Wnt/β-catenin targets by two previous systemic ChIP analyses (39, 40) as well as EBP50 binding sites by our ChIP-on-chip assay. **This gene, although not demonstrated to be Wnt/β-catenin regulated with vigorous experimental evidence, was revealed by previous systemic analysis (39, 40) and this study.