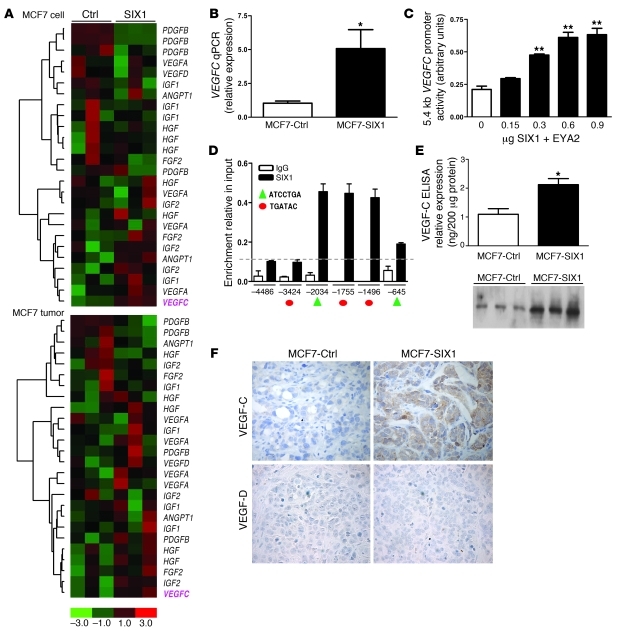
Figure 2. SIX1 transcriptionally activates VEGFC.
(A) Microarray analysis demonstrates that SIX1 overexpression leads to increased VEGFC mRNA in both MCF7-SIX1 cells and MCF7-SIX1 tumors. (B) Quantitation of VEGFC gene expression in MCF7-Ctrl and MCF7-SIX1 using real-time PCR. (C) SIX1 induces VEGFC promoter activity. VEGFC promoter-luciferase reporter constructs were transiently transfected into MCF7 cells, along with increasing amounts of SIX1 and a constant amount of the SIX1 cofactor, EYA2. Luciferase activity was analyzed after 48 hours. (D) ChIP was performed to detect SIX1 presence on the VEGFC promoter in MCF7 cells transfected with SIX1 and EYA2. Protein-DNA complexes were precipitated with a SIX1-specific antibody as well as a control rabbit IgG antibody, after which real-time PCR was performed with primers that flank 5 predicted SIX1 binding sites within the VEGFC promoter (red circles denote the TGATAC binding sites; green triangles denote the ATCCTGA binding sites) as well as 1 upstream region with no predicted SIX1 binding site as a negative control. Dashed line indicates the background non-specific binding of SIX1 and x axis units are base pairs upstream of transcription start site. (E) Functional VEGF-C secreted by MCF7-Ctrl and MCF7-SIX1 cells was measured by ELISA and Western blot analysis (3 clonal isolates of MCF7-Ctrl cells [lanes 1–3] and 3 clonal isolates of MCF7-SIX1 cells [lanes 4–6]) in conditioned medium after serum starvation for 48 hours. (F) MCF7-SIX1 tumors express higher levels of VEGF-C in vivo compared with the MCF7-Ctrl tumors. Immunostaining was used to detect VEGF-C and VEGF-D. Original magnification, ×400. *P < 0.05; **P < 0.01.