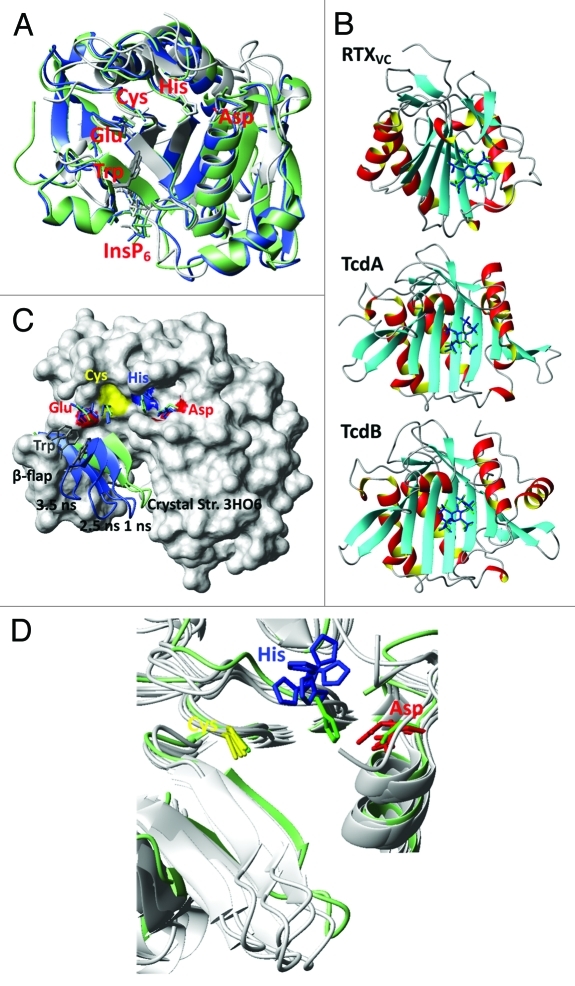
Figure 2. (A) The β-strand core region, the active site and the InsP6 binding site together with the β-flap are conserved and align well for CPD’s of RTXVC (gray), TcdA (green) and TcdB (blue). Shown are the crystal structures with the PDB-codes 3EEB (RTXVC), 3OH6 (TcdA) and 3PA8 (TcdB). They all have InsP6 bound and the substrate is cleaved. Therefore, there are no peptides bound in the active site (we refer to these structures as short). In TcdB the shown residues represent: C698, H653, D587, E743 and W761. (B) Re-docking of InsP6 to the crystal structures of RTXVC (3EEB), TcdA (3HO6) and TcdB (3PA8) with Autodock (blue) shows excellent agreement with the original position of InsP6 (green). (C) β-flap movement of the InsP6 free TcdA CPD (short form 3HO6) during a MD-simulation. Shown residues are C700, H655, D589, E745 and W763. (D) Along with the β-flap the Asp (D589) and His (H655) in the catalytic triad move drastically during an InsP6 free MD-simulation of TcdA CPD (3HO6). Different snapshots at 3.5, 4.0, 5.0, 5.5 and 7.0 ns are compared with the crystal structure positions (green).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.