Abstract
Phylogenetic, genomic and functional analyses have allowed the identification of a new class of putative heme peroxidases, so called APx-R (APx-Related). These new class, mainly present in the green lineage (including green algae and land plants), can also be detected in other unicellular chloroplastic organisms. Except for recent polyploid organisms, only single-copy of APx-R gene was detected in each genome, suggesting that the majority of the APx-R extra-copies were lost after chromosomal or segmental duplications. In a similar way, most APx-R co-expressed genes in Arabidopsis genome do not have conserved extra-copies after chromosomal duplications and are predicted to be localized in organelles, as are the APx-R. The member of this gene network can be considered as unique gene, well conserved through the evolution due to a strong negative selection pressure and a low evolution rate.
Keywords: ascorbate peroxidase, gene duplication, selection pressure, single-copy gene
Ascorbate peroxidases (APx) belong to the class I peroxidase. They have been detected in all chloroplastic containing organisms in which they form a small multigenic family in green lineage.1 They were subjected to some species specific duplications which produced punctual variation in a number of isoforms from 3 to 10. These duplications are probably associated with subfunctionalization. Indeed three major subclasses are defined, based on their cellular localizations: cytoplasmic, peroxisomal and chloroplastic/mitochondrial.2 Recently, an additional group of sequences closely related to APx has been characterized and named ascorbate peroxidase-related (APx-R). Noteworthy, this new class does not seem to be subjected to functional duplication.3
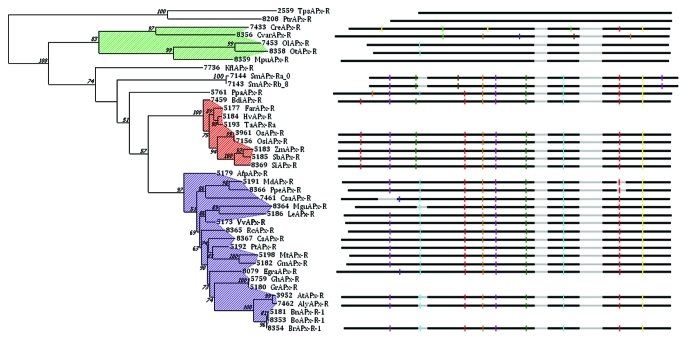
Exhaustive datamining of multiple sequence resources have been performed with available genome and EST libraries to confirm the previous observations. No functional APx-R gene duplication has been detected. Duplicated APx-R are only observed in polyploid organisms: Triticum aestivum, an allohexapolyploid, possesses 3 APx-R,4 and Brassica napus, an allotetraploid, contains at least two independent expressed APx-R with no evidence of conservation of all expected paralogs sequences. Glycine max, an ancient polyploid (palaeopolyploid, tetraploid)5 possesses a single APx-R sequence and one pseudogene while most of APx genes have been detected in duplicated forms. Exhaustive data mining shows that APx-R genes are present in green algae (Chlorophyceae such as Chlamydomonas reinhardtii and Charaphyceae such as Klebsormidium flaccidum) and streptophytes, even though two marginal presences have been detected in chloroplastic diatoms (Table 1). APx-R sequence can be considered as good functional molecular marker because APx-R phylogenetic tree and taxonomic tree are congruent (Fig. 1). More genomic data are needed to determine if all APx-R sequences share the same ancestral sequence or if APx-R from diatoms resulted from a convergent evolution.
Table 1. Ascorbate peroxidase-related (APx-R)-encoding genes identified in different plant species. Exhaustive data mining was performed with all available resources (JGI, NCBI, Phytozome…). When available, EST count and intron number were determined and included in the 5th and 6th columns.
| Name | Taxonomic group | Organism | Sequence Status | Expression (EST count) | Intron number |
|---|---|---|---|---|---|
| PtrAPx-R |
Bacillariophyta (diatoms) |
Phaeodactylum tricornutum |
complete |
4/133887 |
0 |
| TpsAPx-R |
Bacillariophyta (diatoms) |
Thalassiosira pseudonana |
complete |
0/61913 |
0 |
| CreAPx-R |
Chlorophyta (green algae) |
Chlamydomonas reinhardtii |
complete |
18/204076 |
6 |
| CvarAPx-R |
Chlorophyta (green algae) |
Chlorella variabilis |
complete |
0/413 |
5 |
| MpuAPx-R |
Chlorophyta (green algae) |
Micromonas pusilla |
complete |
no |
0 |
| OlAPx-R |
Chlorophyta (green algae) |
Ostreococcus lucimarinus |
complete |
0/17592 |
1 |
| OtAPx-R |
Chlorophyta (green algae) |
Ostreococcus tauri |
complete |
no |
1 |
| VcaAPx-R |
Chlorophyta (green algae) |
Volvox carteri |
partial |
/132038 |
|
| KflAPx-R |
Other Streptophyta |
Klebsormidium flaccidum |
complete |
* |
na |
| AcvAPx-R |
Cryptogam |
Adiantum capillus-veneris |
partial |
1/30540 |
na |
| MpAPx-R |
Cryptogam |
Marchantia polymorpha |
partial |
1/33692 |
na |
| PpaAPx-R |
Cryptogam |
Physcomitrella patens |
complete |
7/362131 |
10 |
| SmAPx-Ra_0 |
Cryptogam |
Selaginella moellendorffii |
complete |
4/93811 |
10 |
| SmAPx-Rb_8 |
Cryptogam |
Selaginella moellendorffii |
complete |
0/93811 |
10 |
| PgAPx-R |
Gymnospermae |
Picea glauca (white spruce) |
partial |
3/313110 |
na |
| PsiAPx-R |
Gymnospermae |
Picea sitchensis (Sitka spruce) |
partial |
1/186637 |
na |
| AmaAPx-R |
Eudicotyledons |
Antirrhinum majus (snapdragon) |
partial |
1/25310 |
na |
| AfpAPx-R |
Eudicotyledons |
Aquilegia formosa x Aquilegia pubescens |
complete |
4/85039 |
na |
| AlyAPx-R |
Eudicotyledons |
Arabidopsis lyrata |
complete |
no |
9 |
| AtAPx-R |
Eudicotyledons |
Arabidopsis thaliana |
complete |
17/1529700 |
9 |
| BnAPx-R-1 |
Eudicotyledons |
Brassica napus (oilseed rape) |
complete |
3/643937 |
na |
| BnAPx-R-2 |
Eudicotyledons |
Brassica napus (oilseed rape) |
partial |
1/643937 |
na |
| BoAPx-R-1 |
Eudicotyledons |
Brassica oleracea (Cauliflower) |
complete |
5/179150 |
na |
| BrAPx-R-1 |
Eudicotyledons |
Brassica rapa |
complete |
0/194305 |
9 |
| CclAPx-R |
Eudicotyledons |
Citrus clementina |
complete |
0/118365 |
9 |
| CsAPx-R |
Eudicotyledons |
Citrus sinensis |
complete |
1/213830 |
9 |
| CsaAPx-R |
Eudicotyledons |
Cucumis sativus |
partial |
0/8128 |
9 |
| EgraAPx-R |
Eudicotyledons |
Eucalyptus grandis |
complete |
0/1910 |
9 |
| EeAPx-R |
Eudicotyledons |
Euphorbia esula |
partial |
1/47543 |
na |
| GmAPx-R |
Eudicotyledons |
Glycine max (soybean) |
complete |
13/1461624 |
9 |
| GmAPx-R[P] |
Eudicotyledons |
Glycine max (soybean) |
pseudogene |
no |
nd |
| GhAPx-R |
Eudicotyledons |
Gossypium hirsutum (cotton) |
complete |
8/273779 |
na |
| GrAPx-R |
Eudicotyledons |
Gossypium raimondii |
complete |
3/63577 |
na |
| HarAPx-R |
Eudicotyledons |
Helianthus argophyllus |
partial |
1/35720 |
na |
| LjAPx-R |
Eudicotyledons |
Lotus japonicus |
partial |
10/242432 |
na |
| LeAPx-R |
Eudicotyledons |
Lycopersicon esculentum (Tomato) |
complete |
6/298289 |
9 |
| MdAPx-R |
Eudicotyledons |
Malus domestica (apple tree) |
complete |
2/324565 |
9 |
| MeAPx-R |
Eudicotyledons |
Manihot esculenta (cassava) |
partial |
1/80681 |
9 |
| MtAPx-R |
Eudicotyledons |
Medicago truncatula (barrel medic) |
complete |
4/269238 |
9 |
| MguAPx-R |
Eudicotyledons |
Mimulus guttatus |
complete |
20/261907 |
8 |
| NtAPx-R |
Eudicotyledons |
Nicotiana tabacum |
partial |
1/332667 |
na |
| PtAPx-R |
Eudicotyledons |
Populus trichocarpa (poplar) |
complete |
1/89943 |
9 |
| PpeAPx-R |
Eudicotyledons |
Prunus persica (peach) |
complete |
0/79584 |
9 |
| RcAPx-R |
Eudicotyledons |
Ricinus communis |
complete |
1/62582 |
9 |
| StAPx-R |
Eudicotyledons |
Solanum tuberosum (Potato) |
partial |
6/249614 |
na |
| ToAPx-R |
Eudicotyledons |
Taraxacum officinale (dandelion) |
partial |
2/41296 |
na |
| VvAPx-R |
Eudicotyledons |
Vitis vinifera (Grape) |
complete |
7/362674 |
9 |
| AGcAPx-R |
Monocotyledons |
Agrostis capillaris |
partial |
1/7743 |
na |
| AsAPx-R |
Monocotyledons |
Avena sativa (Oat) |
partial |
1/25344 |
na |
| BdiAPx-R |
Monocotyledons |
Brachypodium distachyon |
complete |
23/128092 |
10 |
| FarAPx-R |
Monocotyledons |
Festuca arundinacea |
complete |
4/63758 |
na |
| HvAPx-R |
Monocotyledons |
Hordeum vulgare (barley) |
complete |
19/525781 |
na |
| OmAPx-R |
Monocotyledons |
Oryza minuta |
partial |
1/5760 |
na |
| OsiAPx-R |
Monocotyledons |
Oryza sativa (indica) |
complete |
?/203447 |
10 |
| OsAPx-R |
Monocotyledons |
Oryza sativa (japonica) |
complete |
?/987318 |
10 |
| ShyAPx-R |
Monocotyledons |
Saccharum hybrid cultivar (sugarcane) |
partial |
3/282809 |
na |
| SiAPx-R |
Monocotyledons |
Setaria italica |
complete |
0/2741 |
10 |
| SbAPx-R |
Monocotyledons |
Sorghum bicolor |
complete |
6/209828 |
10 |
| TaAPx-Ra |
Monocotyledons |
Triticum aestivum (bread wheat) |
complete |
4/1071453 |
na |
| TaAPx-Rb |
Monocotyledons |
Triticum aestivum (bread wheat) |
partial |
2/1071453 |
na |
| TaAPx-Rd |
Monocotyledons |
Triticum aestivum (bread wheat) |
partial |
2/1071453 |
na |
| ZmAPx-R | Monocotyledons | Zea mays | complete | 16/2019105 | 10 |
no: no EST was found; nd: gene structure cannot be determined; na: no genomic sequence available; * sequence kindly provided by R.Timme.
Figure 1.
Phylogenetic analyses of APx-R were conducted using complete protein sequences from 39 organisms (Gene accession numbers and name are given as PeroxiBase, peroxibase.toulouse.inra.fr/9). Alignment was performed with MAFFT10 and gap columns were discarded. Phylogenetic analysis was conducted with PhyML, using JTT substitution matrix (4), and among-site rate variation was modeled using a Gamma distribution plus a percent of invariant sites and observed frequencies, as advised by ProtTest (JTT+I+G+F). Branch support was calculated by approximate Likelihood Ratio Test (aLTR), and displayed when higher than 50%. Intron positions relatively to the protein alignment are shown, as obtained with CIWOG11 and were included in front of each branch. The hatched areas in green, red and purple stand for green algae, monocots and dicots respectively.
The search performed in EST libraries demonstrated that APx-R are poorly or not expressed in all analyzed organisms with an expression average of 0.003%. Expression analysis in Arabidopsis thaliana with Genevestigator6 confirmed the low level of expression.
In addition to the absence of conserved duplication, high level of sequence conservation is detected (minimum of 50% identity between green algae and streptophyte, and 40% between chloroplastic diatoms and streptophytes). High variability intron positions and number is observed in diatoms and green algae (Fig. 1). However, intron positions and number are highly conserved in higher plants. Only low conservation of the gene structure is observed in the 5′end of the sequences which coincides with the variability of the coding sequence.
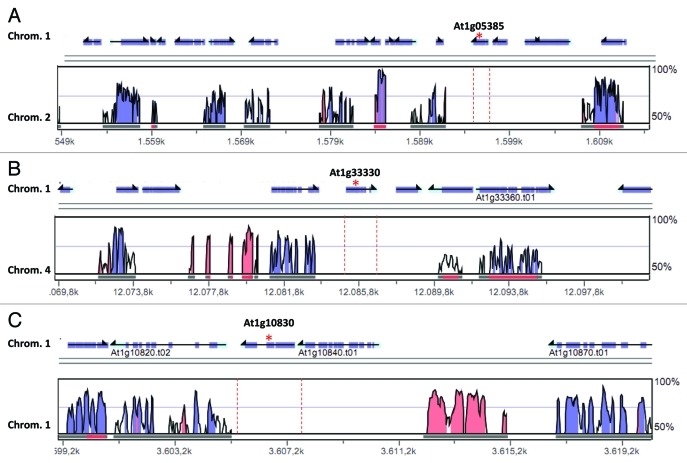
Detailed analysis of Arabidopsis thaliana APx-R co-expression network demonstrated that among the 42 genes listed, 31 encode proteins that are predicted to be localized in organelles, in most cases chloroplasts. These proteins display a great variety of biological functions, but a considerable number of them are implicated in chloroplasts protection against photooxidative damage, which suggests that APx-R could play a role in this protective mechanism as well. Interestingly, more than half of those genes are present as single-copy or as low-copy number in Arabidopsis thaliana (24 among the 42 genes, Table 2), but also in Oryza sativa, Populus trichocarpa and Vitis vinifera genomes. This data confirms that plant proteins predicted to be targeted to organelles are more likely single-copy than expected by chance.7 This could happen because these proteins, when present in the organelles, interact with proteins that are encoded by the organellar genome. In this case, the level of nuclear genome encoded proteins has to be very well controlled inside the cell, so the interaction network will not be disturbed. Looking specifically to the network genes that are single-copy in the specified genomes, we noticed that the majority of the extra-copies of these genes were lost after chromosomal duplications, in a situation very similar to APx-R gene. Thus, it is possible to infer that a great number of single and low-copy genes in this co-expression network could reflect a dose-dependent system, where a raise in copy numbers of such genes would not be favorable to the network. In the Figure 2, LPA19 (At1g05385), peptide release factor (At1g33330) and 15-cis-zeta-carotene isomerase (At1g10830) genes were used as examples. The chromosomal segments that contain these genes in Arabidopsis were duplicated during the evolution and genomic analyses showed that the extra copies were lost during this process (red dashed lines).
Table 2. Co-The list of APx- R co-expressed genes was obtained through the network generated with ATTED-II ver. 6.0 (http://atted.jp/). The putative subcellular localization was predicted through TargetP ver. 1.1 (www.cbs.dtu.dk/services/TargetP/) and Psort ver. 3.0 (www.psort.org/psortb/) and from TAIR databases (www.arabidopsis.org/). The number of copies of each gene was estimated from the data published by Duarte et al., 2010, which listed single and low copy genes in Oryza sativa, Vitis vinifera, Populus trichocarpa and Arabidopsis thaliana genomes.
| Gene | Annotation | Subcellular Localization |
Single-copy gene* | Low-copy gene** | ||
|---|---|---|---|---|---|---|
| TargetP | Psort | TAIR | ||||
| At1g05385 |
LOW PSII ACCUMULATION 19 (LPA19) |
Chlo |
Chlo |
chloroplast, chloroplast thylakoid lumen |
Yes |
- |
| At1g08550 |
NON-PHOTOCHEMICAL QUENCHING 1 (NPQ1); ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1 (AVDE1) |
Other |
Cyto |
chloroplast photosystem II, chloroplast thylakoid lumen |
No |
Yes |
| At1g10830 |
15-CIS-ZETA-CAROTENE ISOMERASE (Z-ISO) |
Chlo |
Chlo |
chloroplast |
Yes |
- |
| At1g27385 |
Unknown protein |
Chlo |
Chlo |
chloroplast |
No |
Yes |
| At1g33290 |
Sporulation protein-related |
Chlo |
Chlo |
n/d |
No |
No |
| At1g33330 |
Peptide chain release factor |
Mito |
Chlo |
chloroplast |
Yes |
- |
| At1g54520 |
Unknown protein |
Chlo |
Chlo |
chloroplast |
Yes |
- |
| At1g64430 |
Unknown protein |
Chlo |
Chlo |
n/d |
No |
Yes |
| At1g67840 |
CHLOROPLAST SENSOR KINASE (CSK) |
Chlo |
Chlo |
chloroplast, chloroplast stroma |
No |
Yes |
| At1g76730 |
5-formyltetrahydrofolate cyclo-ligase family protein |
Chlo |
Chlo |
chloroplast |
No |
Yes |
| At1g78140 |
Methyltransferase-related protein |
Mito |
Chlo |
chloroplast, plastoglobule |
No |
No |
| At1g78995 |
Unknown protein |
Chlo |
Chlo |
n/d |
No |
Yes |
| At2g01620 |
MATERNAL EFFECT EMBRYO ARREST 11 (MEE11) |
Other |
Chlo |
n/d |
No |
No |
| At2g03390 |
uvrB/uvrC motif-containing protein |
Chlo |
Chlo |
chloroplast |
No |
No |
| At2g20860 |
LIPOIC ACID SYNTHASE 1 (LIP1) |
Mito |
Chlo |
mitochondrial matrix, mitochondrion |
No |
No |
| At2g30170 |
Unknown protein |
Chlo |
Chlo |
chloroplast |
No |
No |
| At2g37920 |
EMBRYO DEFECTIVE 1513 (emb1513) |
Chlo |
Chlo |
n/d |
No |
Yes |
| At2g38270 |
CAX-INTERACTING PROTEIN 2 (CXIP2); GLUTAREDOXIN (ATGRX2) |
Chlo |
Chlo |
chloroplast, chloroplast stroma |
Yes |
- |
| At3g10970 |
Haloacid dehalogenase-like hydrolase family protein |
Chlo |
Chlo |
chloroplast |
Yes |
- |
| At3g48560 |
CHLORSULFURON/IMIDAZOLINONE RESISTANT 1 (CSR1); ACETOLACTATE SYNTHASE (ALS); ACETOHYDROXY ACID SYNTHASE (AHAS); TRIAZOLOPYRIMIDINE RESISTANT 5 (TZP5); IMIDAZOLE RESISTANT 1 (IMR1) |
Chlo |
Chlo |
chloroplast |
No |
No |
| At3g53920 |
RNA POLYMERASE SIGMA-SUBUNIT C (SIGC); SIGMA FACTOR 3 (SIG3) |
Chlo |
Chlo |
chloroplast |
No |
No |
| At3g55630 |
A. THALIANA DHFS-FPGS HOMOLOG D (ATDFD) |
Other |
Cyto |
cytosol |
No |
No |
| At4g02260 |
RELA-SPOT HOMOLOG 1 (RSH1); RELA-SPOT HOMOLOG 1 (AT-RSH1); RELA/SPOT HOMOLOG 1 (ATRSH1) |
Chlo |
Plast |
chloroplast |
No |
No |
| At4g10000 |
Electron carrier protein; disulfide oxidoreductase |
Chlo |
Chlo |
chloroplast |
Yes |
- |
| At4g25650 |
ACD1-LIKE (ACD1-LIKE); PROTOCHLOROPHYLLIDE-DEPENDENT TRANSLOCON COMPONENT 52 KDA (PTC52) |
Chlo |
Plast |
chloroplast, chloroplast envelope |
No |
No |
| At4g27600 |
NECESSARY FOR THE ACHIEVEMENT OF RUBISCO ACCUMULATION 5 (NARA5) |
Chlo |
Chlo |
chloroplast |
Yes |
- |
| At4g30310 |
Ribitol kinase protein |
Other |
Chlo |
chloroplast |
No |
No |
| At4g32320 |
ASCORBATE PEROXIDASE-RELATED (APX-R) |
Chlo |
Chlo |
cytosol |
Yes |
- |
| At4g33630 |
EXECUTER1 (EX1) |
Chlo |
Chlo |
thylakoid membrane |
No |
No |
| At5g02250 |
EMBRYO DEFECTIVE 2730 (EMB2730); RIBONUCLEOTIDE REDUCTASE 1 (RNR1); ARABIDOPSIS THALIANA MITOCHONDRIAL RNASE II (ATMTRNASEII) |
Chlo |
Chlo |
chloroplast, mitochondrion |
Yes |
- |
| At5g03900 |
Unknown protein |
Chlo |
Plast |
chloroplast envelope |
Yes |
- |
| At5g04360 |
PULLULANASE 1 (ATPU1); LIMIT DEXTRINASE (ATLDA); PULLULANASE 1 (PU1) |
Chlo |
Chlo |
chloroplast |
No |
Yes |
| At5g06340 |
ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 27 (ATNUDX27) |
Chlo |
Chlo |
chloroplast |
No |
No |
| At5g08340 |
Riboflavin biosynthesis protein-related |
Other |
Chlo |
cellular_component unknown |
No |
No |
| At5g08410 |
FERREDOXIN/THIOREDOXIN REDUCTASE SUBUNIT A2 (FTRA2) |
Chlo |
Chlo |
chloroplast |
No |
Yes |
| At5g13720 |
Unknown protein |
Chlo |
Plast |
chloroplast, chloroplast inner membrane, chloroplast envelope |
No |
No |
| At5g18140 |
DNAJ heat shock N-terminal domain-containing protein |
Chlo |
Nuclear |
n/d |
No |
No |
| At5g19540 |
Unknown protein |
Chlo |
Chlo |
chloroplast |
No |
Yes |
| At5g26820 |
MULTIPLE ANTIBIOTIC RESISTANCE 1 (MAR1); IRON REGULATED 3 (IREG3) |
Chlo |
Plast |
chloroplast, chloroplast envelope |
No |
Yes |
| At5g38510 |
Rhomboid family protein |
Chlo |
Nuclear |
integral to membrane |
Yes*** |
- |
| At5g57040 |
Lactoylglutathione lyase family protein |
Chlo |
Chlo |
chloroplast |
Yes |
- |
| At5g65685 | Soluble glycogen synthase-related protein | Chlo | Chlo | chloroplast | No | No |
Single-copy genes in Oryza sativa, Vitis vinifera, Populus trichocarpa and Arabidopsis thaliana genomes, according to Duarte et al., 2010.7 **Genes present as one or two copies in at least one of the analyzed genomes. ***Not present in Oryza sativa.
Figure 2.
Comparative analyses between the genomic regions containing APx-R co-expressed genes At1g05385 (A), At1g33330 (B) and At1g10830 (C) and the DNA segments generated after these regions were duplicated. Peaks correspond to conserved DNA sequences (exons, in most cases) in the Arabidopsis duplicated genomic regions. Horizontal arrows above the graphics represent the annotated genes in Arabidopsis genome (AGI codes are shown). The red dot lines indicate the absence of the APx-R co-expressed gene in duplicated region. The VISTA program (www-gsd.lbl.gov/vista/) was used in these analyses.
The hypothesis of conserved unique genes has already been proposed.8 However further analyses are mandatory to precisely evaluate the extension of the proposal of a complex network of unique gene, taking into consideration that many other neighbor genes were also deleted from these genomic regions. The conservation of this unique gene network indicates that they are under a strong negative selection pressure and subjected to low evolution rate.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18098
References
- 1.Passardi F, Bakalovic N, Teixeira FK, Margis-Pinheiro M, Penel C, Dunand C. Prokaryotic origins of the non-animal peroxidase superfamily and organelle-mediated transmission to eukaryotes. Genomics. 2007;89:567–79. doi: 10.1016/j.ygeno.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Teixeira FK, Menezes-Benavente L, Galvao VC, Margis R, Margis-Pinheiro M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta. 2006;224:300–14. doi: 10.1007/s00425-005-0214-8. [DOI] [PubMed] [Google Scholar]
- 3.Lazzarotto F, Teixeira FK, Rosa SB, Dunand C, Fernandes C, Fontenele AD, et al. Ascorbate peroxidase-related (APx-R) is a new heme-containing protein functionally associated with ascorbate peroxidase but evolutionarily divergent. New Phytol. 2011;191:234–50. doi: 10.1111/j.1469-8137.2011.03659.x. [DOI] [PubMed] [Google Scholar]
- 4.Kerby K, Kuspira J. The phylogeny of the polyploid wheats Triticum aestivum (bread wheat) and Triticum turgidum (macaroni wheat) Genome. 1987;29:722–37. doi: 10.1139/g87-124. [DOI] [Google Scholar]
- 5.Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 6.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duarte JM, Wall PK, Edger PP, Landherr LL, Ma H, Pires JC, et al. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol. 2010;10:61. doi: 10.1186/1471-2148-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armisén D, Lecharny A, Aubourg S. Unique genes in plants: specificities and conserved features throughout evolution. BMC Evol Biol. 2008;8:280. doi: 10.1186/1471-2148-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koua D, Cerutti L, Falquet L, Sigrist CJA, Theiler G, Hulo N, et al. PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res. 2009;37:D261–6. doi: 10.1093/nar/gkn680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–98. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 11.Wilkerson MD, Ru YB, Brendel VP. Common introns within orthologous genes: software and application to plants. Brief Bioinform. 2009;10:631–44. doi: 10.1093/bib/bbp051. [DOI] [PubMed] [Google Scholar]