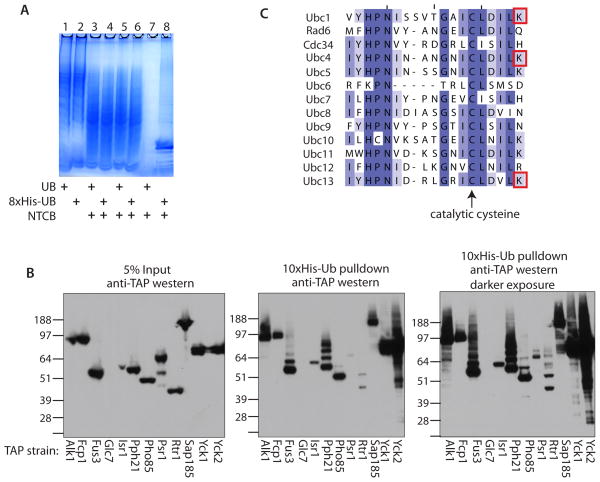
Figure 2.
(A) Analysis of NTCB treatment and his-tag purifications of protein lysates from yeast strains either expressing 8xHis-ubiquitin or untagged ubiquitin by SDS-PAGE and coomassie staining. Lanes 1 and 2- whole cell lysates; lanes 3 and 4- protein fragments after treatment with NTCB; lanes 5 and 6- protein fragments post dialysis; lanes 7 and 8- metal affinity purified Ub-fragments. Yeast strains as indicated. (B) Validation of ubiquitinated protein kinases and phosphatases. The TAP strain for each of the indicated kinases and phosphatases was transformed with a 10xHis-ubiquitin expression plasmid and grown to mid-log phase. The yeast were lysed in denaturing buffer. 5% of the lysate was analyzed by anti-TAP western blot (left panel) and the 10xHis-ubiquitinated proteins were purified from the remaining lysate using metal affinity resin. The resin was boiled in sample buffer + ∀-mercaptoethanol and analyzed by anti-TAP western blot (middle panel; darker exposure in the right panel). Molecular weights are indicated. (C) The 13 yeast E2 ubiquitin conjugating enzymes aligned by ClustalW and colored according to percent identity, darker purple = higher conservation. The catalytic cysteine is highlighted with an arrow and the red boxes indicate the lysines that were identified as having a ubiquitin remnant mass shift in this study.