Fig. 2.
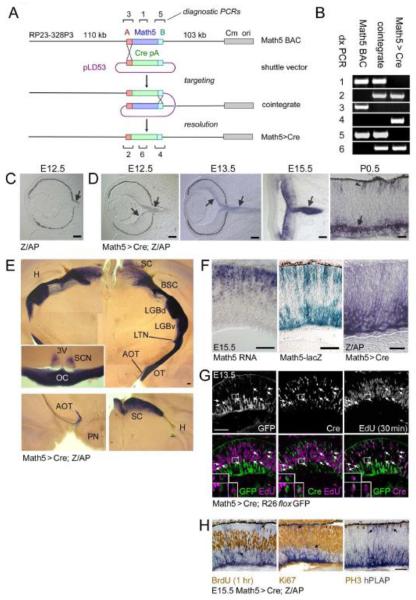
Construction and expression of the Math5>Cre transgene. (A) Math5>Cre BACs were generated in E. coli by a two-step homologous recombination procedure. The single-exon Math5 open reading frame was precisely replaced with a nlsCre-pA cassette, using “A” and “B” homology arms derived from 5′ (red box) and 3′ (cyan box) UTRs. The pLD53 shuttle vector contains recA recombinase, positive (amp) and negative (sacB) selection cassettes, and the R6K origin of replication. The pBACe3.6 vector (gray box) contains the chloramphenicol resistance gene (Cm) and P1 origin (ori). (B) Confirmation of recombinant BAC structure using diagnostic (dx) PCRs 1-6 indicated in panel A (assembled from multiple gels). (C-F) Developmental expression pattern. Math5>Cre mice were crossed to mice carrying the Z/AP transgene, which permanently reports Cre activity. Alkaline phosphatase (hPLAP)-positive RGCs (purple, arrows) are first observed at E12.5 in double transgenic embryos (D), while control littermates (C) containing only the Z/AP transgene are negative. hPLAP activity increases from E13.5 to E15.5 as RGCs develop and form the optic nerve. By P0.5, hPLAP activity is abundant in RGCs (arrow) and can be detected in some photoreceptors (arrowhead); however most of the retina is unlabeled. (E) Composite images of 250 μm coronal vibratome sections through the adult thalamus and optic chiasm (inset) show the axonal projections of RGCs derived from Math5+ precursors. Lower panels show sections through the accesory optic system (left) and superior colliculus (right). Labeled RGCs project to all major ganglion cell target sites in the CNS. No significant staining was observed in the cerebral cortex or hippocampus. (F) Kinetics of Math5 expression in the E15.5 retina. Math5 mRNA (in situ hybridization) is expressed in retinal progenitor cells, while the cytoplasmic Math5-lacZ knock-in allele labels progenitors (sclerad) and developing RGCs (vitread), which have recently transcribed Math5 (β-galactosidase activity). hPLAP activity in Math5>Cre; Z/AP mice is localized to developing RGCs on the vitread side of the retinal epithelium. These patterns demonstrate the spatiotemporal progression of Math5 expression, if one considers the perdurance of β-galactosidase, the delay associated with Cre excision, and interkinetic nuclear migration. Math5 is expressed transiently in progenitors that become RGCs. (G-H) Math5>Cre retinas co-stained for lineage tracers and cell cycle markers. Cre is expressed with the same kinetics as Math5, whereas GFP or hPLAP reporters are expressed with a delay. (G) At E13.5, Cre+ EdU+ cells are present (30 min chase, arrows, 33 of 394 Cre+ cells), but no GFP+ EdU+ cells are observed (0 of 309 GFP+ cells). (H) At E15.5, no hPLAP+ cells (arrows) are co-labeled with BrdU (1 hr chase), PH3, or Ki67 (marks late G1 through M phase). Together, these results indicate that cells in the Math5 lineage do not re-enter the cell cycle. pA, polyadenylation signal; SC, superior colliculus; BSC, brachium of the superior colliculus; LGBd, lateral geniculate body, pars dorsalis; LGBv, lateral geniculate body, pars ventralis; LTN, lateral terminal nucleus; AOT, accessory optic tract; OT, optic tract; TV, third ventricle; SCN, suprachiasmatic nucleus; OC, optic chiasm; H, hippocampus; PN, pons. Scale bars, 100 m in C-F; 50 m in G-H.