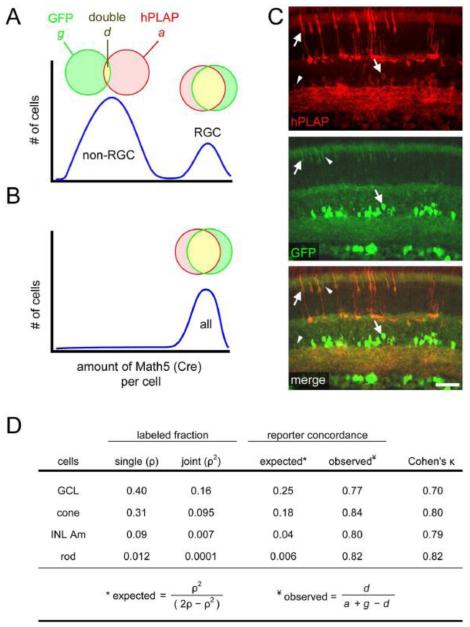
Fig. 4.
All Math5>Cre progenitors express similar levels of Cre, regardless of cell fate. Math5>Cre lineage analysis was performed using Z/AP and R26floxGFP reporters simultaneously, to evaluate the heterogeneity of Math5 expression among progenitors. This analysis assumes that the probability of reporter activation in a given cell is determined by the cumulative amount of Cre recombinase expressed by that cell. (A-B) Two models for Math5 (Cre) expression. (A) Bimodal expression. In this model, Math5+ progenitors giving rise to non-ganglion cell types express Cre weakly (left peak), so reporter activation in these cells is inefficient, and consequently few of their descendants co-express GFP and hPLAP. RGCs in the same retinas express Cre strongly (right peak) and are expected to have high concordance. (B) Uniform expression. In this model, every Math5+ progenitor expresses Cre strongly, so concordance is very high for all cell types (B, right). (C) Retinas of adult Math5>Cre; Z/AP; R26floxGFP mice immunostained for hPLAP and GFP. Double-labeled cells (arrows) greatly outnumber single-labeled cells (arrowheads). (D) The observed concordance between hPLAP and GFP reporters was ~80%, which is significantly greater than expected by chance (Cohen’s κ > 0.7). This value was similar for all cell types, indicating that Math5 is expressed at uniform levels by a subpopulation of progenitor cells, only some of which develop as RGCs. The labeled fractions (ρ) are based on data in Table 1. GCL includes RGCs and displaced amacrines; INL Am, inner nuclear layer amacrine. Scale bar, 50 m.