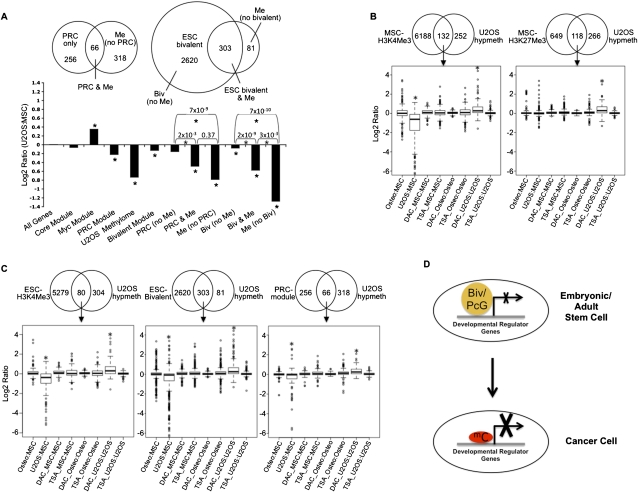
Figure 6.
PRC-module and ESC-bivalent genes undergo significant repression of expression upon hypermethylation in cancer. (A, top panel) Venn plot of the overlap between the 384 genes hypermethylated in U2OS and the PRC module (Kim et al. 2010) (left) or the ESC-bivalent module (right). (Bottom panel) Expression changes between U2OS and MSCs for the Myc, Core, PRC-modules, ESC-bivalent genes, and the set of genes hypermethylated in osteosarcoma (termed “U2OS Methylome”). (*) Significant change from background based on Wilcoxon rank-sum test. The methylated genes that are not bivalent in ESCs (called “Me no bivalent”) are all marked by H3K4Me3. (B) Effect of 5-deoxy-aza-cytidine (DAC) and trichostatin (TSA) on re-expression of U2OS-hypermethylated genes marked by H3K4Me3 or H3K27Me3 in MSCs (B) or those marked by H3K4Me3, bivalent or PRC in ESCs (C). The log2 gene expression ratios for these genes from Agilent gene expression data summarizing the relative expression of osteoblasts versus MSCs, U2OS versus MSCs, and the relative expression of DAC- or TSA-treated MSCs, osteoblasts, and U2OS versus their control-treated counterparts are shown in B and C. (*) P-values < 0.001. (D) Model depicting the change of repressive mechanism from bivalent/PcG-marking to DNA hypermethylation. Developmental regulator genes are marked by bivalent (embryonic/adult stem cell) or PcG (adult stem/early progenitor cells) and have relatively low levels of expression. In tumors, DNA hypermethylation (mC) of bivalent/PcG-marked genes in stem cells leads to tight silencing at these genes. This epigenetic switch is responsible for the stable silencing seen at developmental regulators in cancer cells and may explain how cancer cells recapitulate aspects of the transcriptional and phenotypic state of stem cells.