Figure 4.
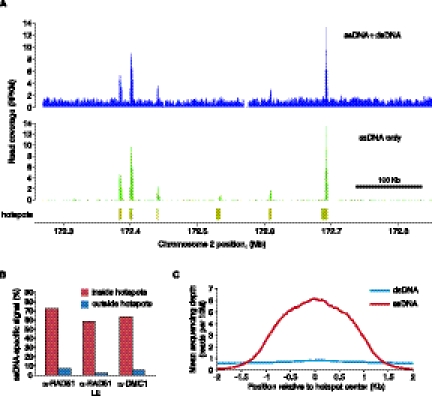
Most of the DSB hotspot signal is ssDNA-specific. (A) ssDNA identification strongly reduces background in low-enrichment ChIP α-RAD51 Psmc3ip−/−LE library. ssDNA + dsDNA or type I ssDNA only coverage profiles (reads per 1 kb, per million reads, RPKM) are plotted for a region on mouse chromosome 2. Coverage profiles were normalized to total number of ssDNA + dsDNA reads. α-RAD51 Psmc3ip−/− LE library was prepared without using kinetic enrichment and ssDNA was computationally identified in the sequencing data. (B) The proportion of ssDNA-specific signal inside or outside all hotspots. While most of the signal inside hotspots is retained after ssDNA identification, outside of the hotspots, the background is reduced 10-fold or more. Sample specificity is plotted for α-DMC1 Psmc3ip−/− (α-DMC1), α-RAD51 Psmc3ip−/− (α-RAD51), and α-RAD51 Psmc3ip−/− LE (α-RAD51 LE) samples. (C) Most of the signal inside hotspots originates from ssDNA. Mean depth of ssDNA and dsDNA read coverage (reads per 10 million reads) across all 9874 published mouse hotspots are plotted in 5-bp increments.