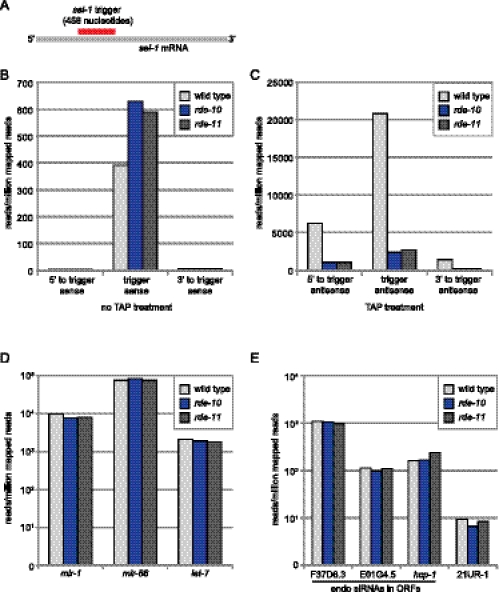
Figure 5.
RDE-10 and RDE-11 are required for secondary siRNA synthesis. (A) Schematic representation of sel-1 RNAi trigger and sel-1 mRNA. (B–E) The mean abundance of small RNAs from two independent populations of wild-type, rde-10(hj20), and rde-11(hj37) animals undergoing sel-1 RNAi is shown. (B) Sense sel-1 siRNAs without TAP treatment. (C) Antisense sel-1 siRNAs with TAP treatment. (D) Selected miRNAs (mir). (E) Selected endogenous siRNAs and one 21U-RNA. Reads per million (RPM) was calculated by (reads aligned to region/all aligned reads for sample) × 1,000,000. A 5′ ligation-dependent method was used in library preparations. Small RNAs with 5′ monophosphate were better represented in libraries without TAP treatment. Small RNAs with 5′ triphosphate were better represented in libraries with TAP treatment.