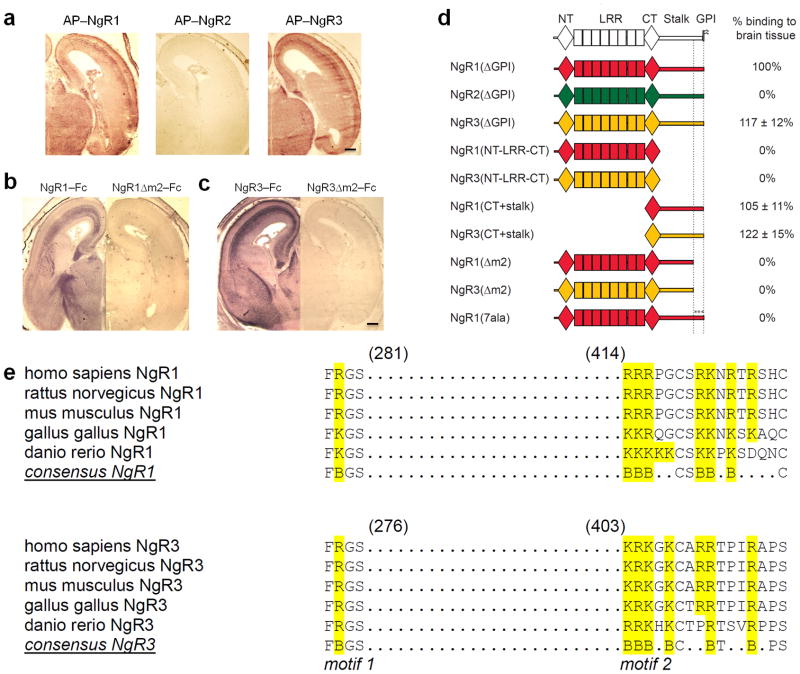
Figure 2. NgR1 and NgR3, but not NgR2, contain two discontinuous and evolutionarily conserved sequence motifs necessary for binding to brain tissue.
(a) Coronal sections of E18 rat brain showing the binding pattern of AP–NgR1 and AP–NgR3. No binding is observed for AP–NgR2. (b–c) Binding of (b) NgR1–Fc and (c) NgR3–Fc to E18 brain sections is abolished upon deletion of a cluster of basic residues (motif m2) in the stalk region. (d) Schematic of receptor deletion constructs and their relative binding to E18 rat brain tissue compared to soluble NgR1 [NgR1(ΔGPI)]. Soluble NgR1 (red) and NgR3 (yellow), but not NgR2 (green), bind strongly to brain tissue sections. The LRRs of NgR1, previously shown to participate in myelin inhibitor binding, are dispensable for binding to neural tissue. Deletion of a cluster of basic amino acid residues in the C–terminal region of the NgR1 and NgR3 stalk (motif m2), or replacement of these residues with alanines [NgR1(7ala)], completely abolishes binding. (e) Sequence alignment of binding motifs m1 and m2 of NgR1 and NgR3. In the LRR–CT domain, residues F278 and R279 in NgR1 and residues F273 and R274 in NgR3 (motif m1) are important for GAG binding. Motif m2 is comprised of a cluster of basic amino acids, including residues 414–426 in NgR1 and residues 403–415 in NgR3. The conserved residues are bolded and the basic residues are in blue. Scale bar, 40μm.