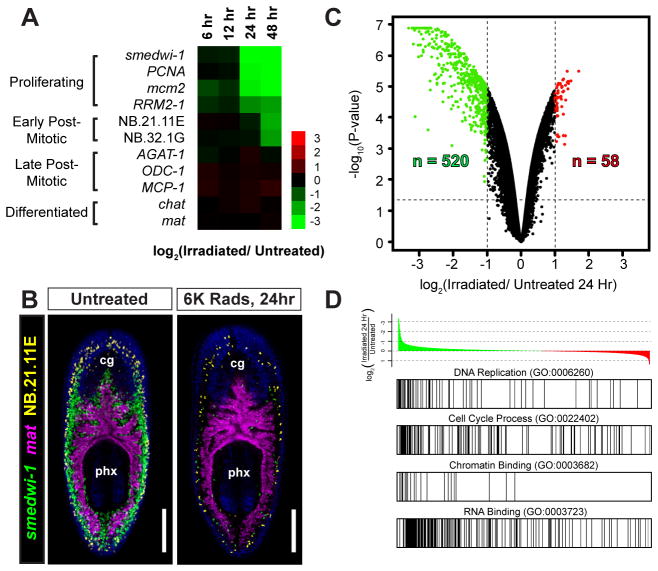
Figure 1. Identification of Irradiation-Sensitive Transcripts in Adult Planarians by Microarrays.
(A) Heat map illustrating mRNA depletion kinetics following 6,000 Rads γirradiation. Markers for proliferating cells (smedwi-1, PCNA, mcm2, RRM2-1), post-mitotic cells (NB.21.11E, NB.32.1G, AGAT-1, ODC-1, and MCP-1), neurons (chat), and intestine (mat) are shown. (B) Whole-mount triple-fluorescence in situ hybridization (FISH) shows the anatomical distribution of proliferative cells (neoblasts) with untreated and 24-hour irradiated animals. Shown are projections through z-stacks of multiple confocal planes in the interior of entire animals. Pharynx (px) and cephalic ganglia (cg) are indicated. Ventral views, anterior up. Scale bars, 200 μm. (C) Volcano plot showing transcripts depleted (green) or upregulated (red) 24 hours after irradiation. See Supplemental Table 1. (D) Gene set enrichment analysis (GSEA) with annotated gene list pre-ranked by log2 ratio (24-hour irradiated/untreated). Example gene sets enriched among irradiation-depleted transcripts are shown. See Supplemental Table 2.