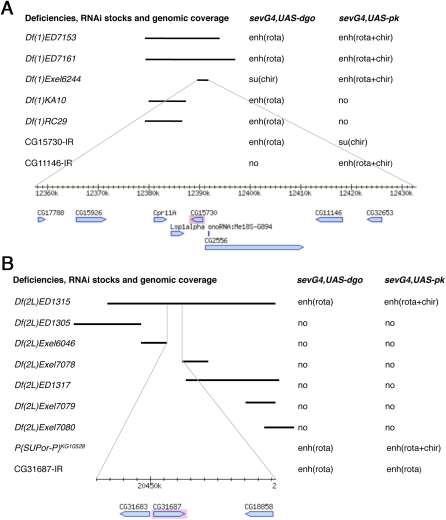
Figure 4 .
Single gene identification strategies for two DrosDel deficiencies, Df(1)ED7161 and Df(2L)ED1315. Deficiencies and their coverage indicated by black bars are shown on the left and their genetic interactions are listed on the right. Fine lines at the bottom of each panel connect to a genomic map for the area that was tested by RNAi’s (based on “MapBrowse, FlyBase”). (A) Mapping several interaction areas in Df(1)ED7161, which enhanced ommatidial rotation defects of sev-GAL4, UAS-dgo and enhanced rotation and chirality defects of sev-GAL4, UAS-pk. Two different genomic areas accounted for these effects, as identified by Df(1)KA10 and Df(1)RC29 for one of the interaction and Df(1)Exel6244 for the other. Df(1)Exel6244 was further studied since it modified both screen genotypes. CG15730 and CG11446 (now fused with CG32653 and called CG42251) were found to reproduce, each a subset of the original genetic interaction. (B) For Df(2L)ED1315, mapping to a single gene was done by exclusion. DrosDel deficiency Df(2L)ED1315 enhanced ommatidal rotation defects in sev-GAL4, UAS-dgo and enhanced both rotation and chirality defects in sev-GAL4, UAS-pk. Several overlapping or subdividing deficiencies were assayed for genetic interaction, but showed no modification of either genotype. This left three genes near the center of Df(2L)ED1315 that were not covered by any of the smaller deficiencies. We therefore tested these three genes and found that RNAi for CG31687 (an APC8 paralog) interacted similarly to Df(2L)ED1315 with the two screening genotypes, as did a P-element insertion KG10528 for CG2508 (APC8/cdc23).