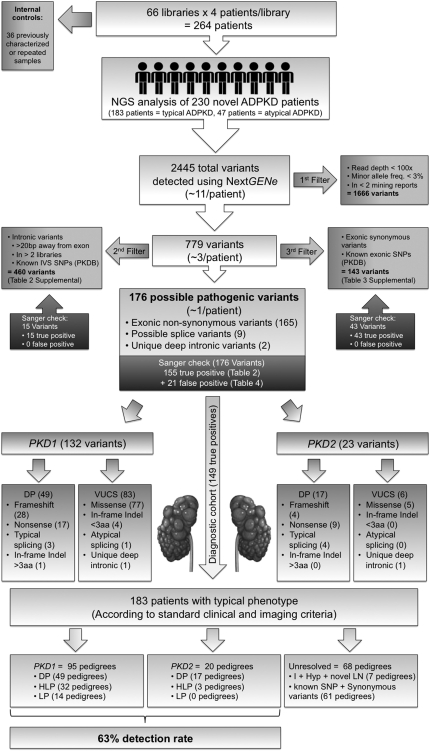
Figure 4.
Schematic diagram illustrating the workflow utilized for filtering, parsing, and re-confirming all of the variants derived after the initial data mining in the discovery experiment (Figure 3B). After read re-alignment and variant calling, quality filtering removed 1666 low-confidence variants from the initial pool of 2445 called variants, resulting in 779 high-confidence variants (see Concise Methods for details). This reduced the average number of variants per patient from approximately 10 to 3. Parsing by likelihood of disease association further removed 460 common intronic variants and 143 synonymous or known nonsynonymous exonic polymorphisms, resulting in 176 possible pathogenic variants (approximately 1 per patient). After Sanger re-confirmation, the 155 true positive variants were classified for pathogenicity either as definitely pathogenic (DP) or VUCS, which were further classified as highly likely pathogenic (HLP), likely pathogenic (LP), indeterminate (I), likely hypomorphic (Hyp), and likely neutral (LN) (Table 2 and see Concise Methods). As we focused on the diagnostic cohort of 183 pedigrees (arrow), the genotypes in the pedigrees from this subgroup were classified based on the most pathogenic mutation found as having a DP genotype (49 PKD1 pedigrees and 17 PKD2 pedigrees), an HLP genotype (32 PKD1 pedigrees and 3 PKD2 pedigrees), an LP genotype (14 PKD1 pedigrees) (Table 2). Of the 68 pedigrees with unresolved genotype from the diagnostic cohort of 183 patients, 7 carried I, Hyp, or novel LN genotypes (Table 2). The remaining 61 pedigrees from the diagnostic cohort of 183 patients only had synonymous or known polymorphisms (not shown). Hence, within the diagnostic cohort of 183 typical ADPKD according to standard clinical and imaging criteria, the 115 of 183 resolved pedigrees accounted for a final detection rate of 63%. DP, definitely pathogenic; HLP, highly likely pathogenic; LP, likely pathogenic; I, indeterminate; Hyp, likely hypomorphic; LN, likely neutral.