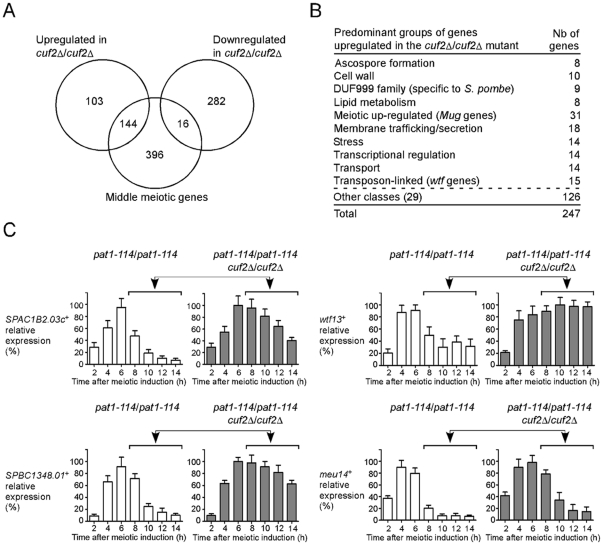
Figure 5. Effect of cuf2Δ/cuf2Δ deletion on the expression of S. pombe genes.
A, Venn diagram representing the numbers of genes with higher and lower expression levels in the cuf2Δ/cuf2Δ disruption strain as compared to the wild-type strain. The diagram also shows the overlap between middle-phase meiotic genes and all differentially expressed genes identified in cuf2Δ/cuf2Δ cells. B, Genes that exhibit higher expression levels in cuf2Δ/cuf2Δ cells were grouped in different functional families. C, pat1-114/pat1-114 (cuf2+/cuf2+) and pat1-114/pat1-114 cuf2Δ/cuf2Δ strains were pre-synchronized by nitrogen starvation and were then induced to undergo synchronous meiosis under basal conditions. At the indicated times following meiotic induction, total RNA was prepared from both the cuf2+/cuf2+ strain and its cuf2Δ/cuf2Δ mutant derivative and was analyzed by RNase protection assay. The histograms shown the quantifications of three independent RNase protection experiments. Ten hours after meiotic induction, the results indicate that the levels of the SPAC1B2.03c+, wtf13+, SPBC1348.01+ and meu14+ transcripts in cuf2Δ/cuf2Δ mutant cells were derepressed above those observed in cuf2+/cuf2+ cells under the same conditions (as indicated with arrows and brackets).