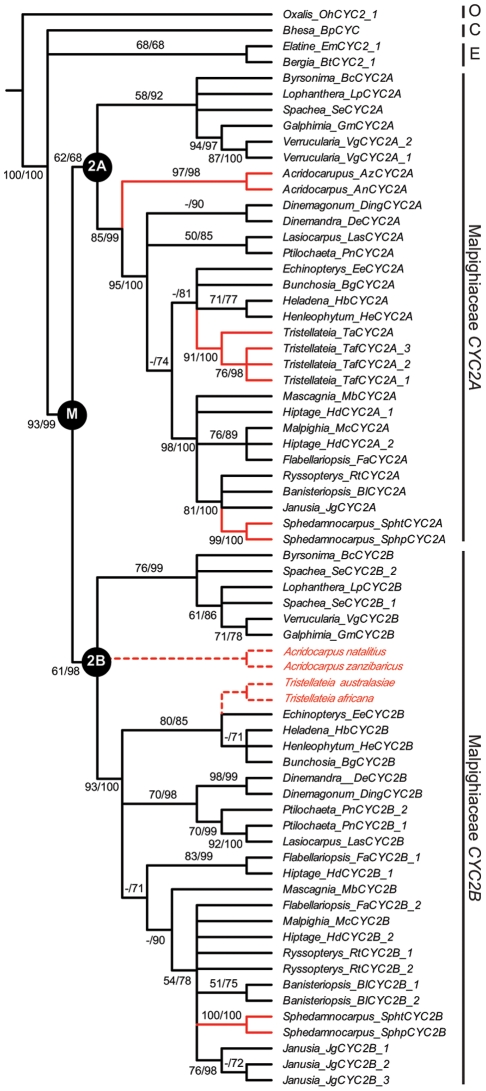
Figure 2. Phylogeny of CYC2-like genes for Malpighiaceae.
Bayesian majority rule consensus topology shown; clades with >50% maximum likelihood (ML) bootstrap support and >60% Bayesian posterior probabilities depicted above lines, respectively. ML bootstrap support <50% indicated with a hyphen. Inferred gene tree is reflective of accepted species tree relationships [40]. Accessions highlighted in red include the three Old World clades examined here that exhibit parallel floral phenotypes–Acridocarpus, African Sphedamnocarpus, and Tristellateia. Accessions labeled with dotted lines signify inferred gene losses in Acridocarpus natalitius, A. zanzibaricus, Tristellateia australasiae, and T. africana. See Supplementary Table S2 for species identities and voucher information. C, Centroplacaceae; E, Elatinaceae; M, Malpighiaceae; O, Oxalidaceae.