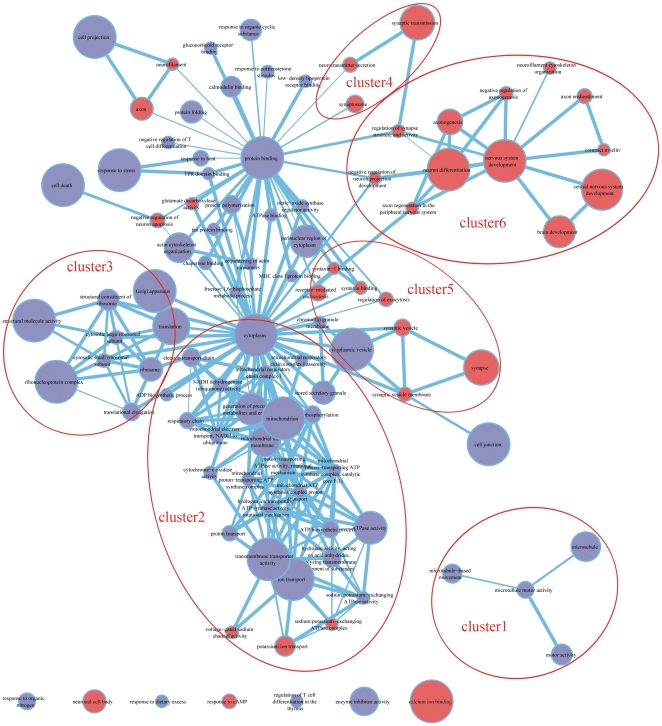
Figure 4. Network of enriched GO terms derived from differentially expressed genes between schizophrenia and control using GOseq software.
The enriched GO terms are organised as a weighted similarity network, where nodes represent enriched GO terms and weighted links between the nodes represent the overlap score calculated from the number of genes two GO terms share. By grouping enriched GO terms into network clusters based on their overlapping extent, 6 distinct functional clusters are identified, which are (1) microtubule related motility, (2) mitochondrial function and ATP production, (3) ribosome and translation activity, (4) neurotransmission related functions, (5) synaptic vesicle trafficking, and (6) neural development. GO terms with more overlapping genes are placed closer together; node size represents the number of genes in the pathway; edge thickness is proportional to the overlap between pathways. Nodes highlighted in pink represent GO terms directly relevant to the pathophysiology of schizophrenia.