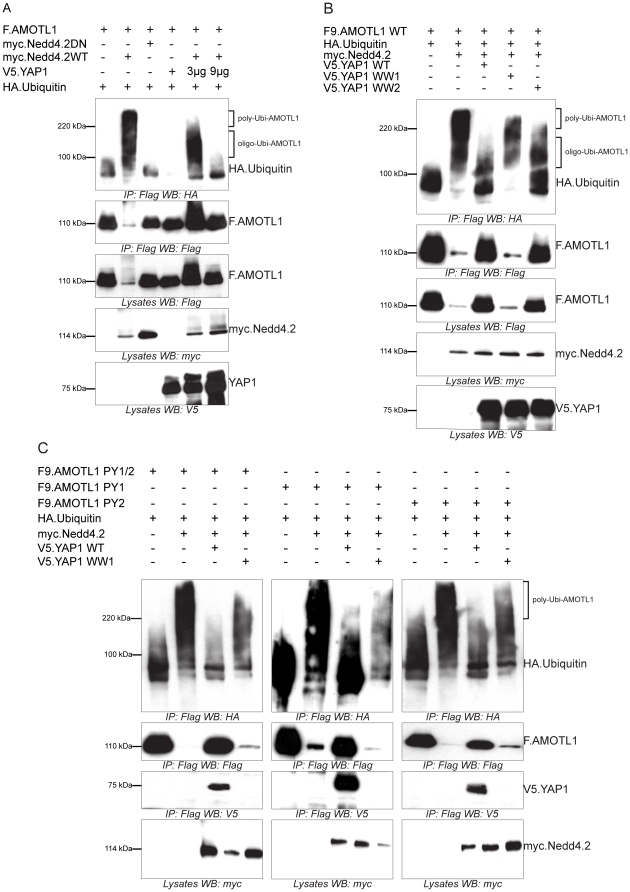
Figure 4. YAP1 inhibits AMOTL1 degradation via its first WW domain.
A, Nedd4.2 facilitates the ubiquitylation of AMOTL1. HEK293T cells were transfected with AMOTL1, Nedd4.2 and YAP1, as indicated. AMOTL1 was precipitated and probed for incorporated HA-tagged ubiquitin. AMOTL1 levels were determined by re-probing the blot with anti-Flag antibody. A catalytically inactive (DN) Nedd4.2 was used as a control for ligase activity. B, Wild type YAP1 and the YAP1WW2 mutant, but not the YAP1WW1 mutant, reduce ubiquitylation of AMOTL1. HA-tagged ubiquitin (HA.Ubiquitin) and expression vectors, as indicated, were co-expressed in HEK293T cells. AMOTL1 was precipitated by M2 sepharose beads, and then probed with antibody to HA to identify ubiquitylated AMOTL1. C, YAP1 protects both single and double AMOTL1 PPxY mutants against Nedd4.2-mediated ubiquitylation. Either of AMOTL1310–313A (F.AMOTL1PY1), AMOTL1367–370A (F.AMOTL1PY2) or AMOTL1310–313A/367–370A (F.AMOTL1PY1/2) were co-expressed with different combinations of HA.Ubiquitin, myc.Nedd4.2, V5.YAP1 and V5.YAP1 WW1, as indicated. Cell extracts were precipitated with M2 beads and blotted with anti-HA antibody. In blots for ubiquitin, brackets indicate molecular weights that correspond to poly- or oligo-ubiquitylation of AMOTL1. Poly-ubiquitylation results in complexes with molecular weights above 220 kDa which signal degradation of the protein by the 26S proteasome. Complexes between 100 kDa and 220 kDa indicate oligo-ubiquitylated peptides that are generally not degraded by the 26S proteasome.