Figure 2. Basis of affinity enhancement for IL-2Rβ from structural and molecular dynamics characterization of D10 super-2.
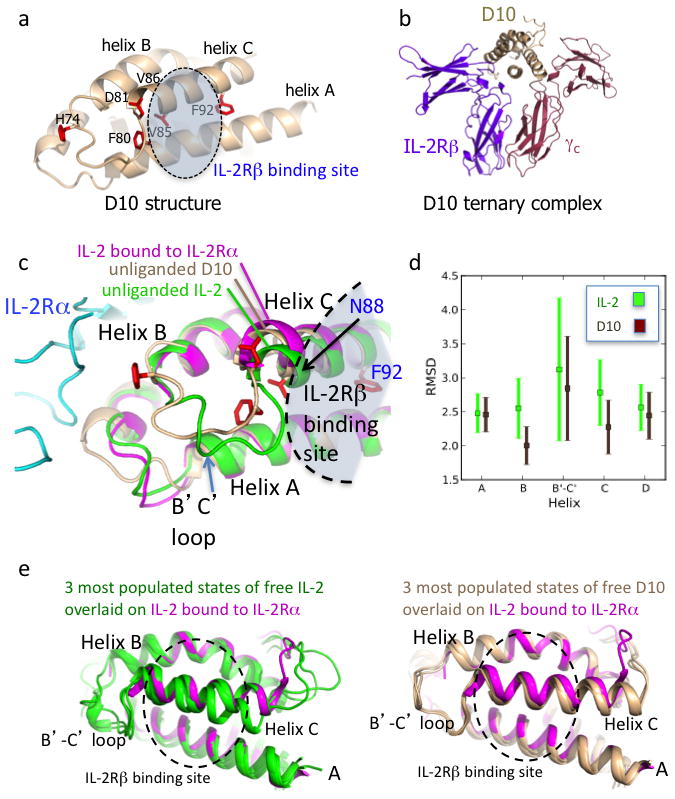
a, Crystal structure of the D10 super-2 at 3.1 Å with mutated residues in red (see also Supplementary Table 1 and Supplementary Fig. 7a). b, D10 in complex with human IL-2Rβ and γc preserves the WT receptor dimer geometry (see also Supplementary Fig. 7b). c, The unliganded D10 super-2 helix C (brown), moves towards its hydrophobic core compared to unliganded WT IL-2 (green, PDBID 3INK). This helix C position is more similar to that of helix C in IL-2 bound to IL-2Rα (purple, PDBID 1Z92) (see also Supplementary Fig. 8). d, A 40ns MD simulation shows a reduction of the average RMSD for the B and C helices, and the B-C loop in D10 versus IL-2 (see also Supplementary Fig. 8c). Error bars represent the standard error of the RMSD. e, Helix C in IL-2 (green, left panel) drifts during the MD simulation more than super-2 D10 (brown, right panel) when compared to IL-2 bound to IL-2Rα (purple).
