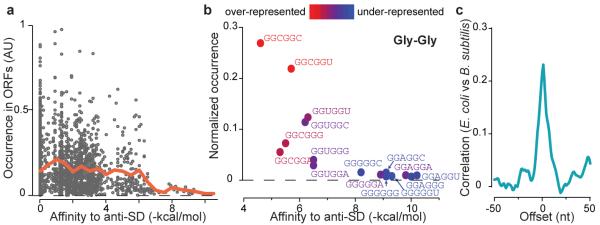
Fig. 4.
Selection against SD-like sequences and the constraint on protein coding. a, Rate of occurrence of hexanucleotide sequences in E. coli messages relative to their affinity to the anti-SD site. The orange line shows the average occurrence within a bin size of 0.5 kcal/mol. b, Occurrence of codon-pairs for glycine-glycine residues relative to their affinity to the anti-SD site. The color coding represents the enrichment in occurrence of codon pairs after correcting for the usage of single codons. c, Cross-correlation function of ribosome occupancy profiles between conserved genes in E. coli and B. subtilis. Zero offset means the two sequences are aligned at each amino acid residue.