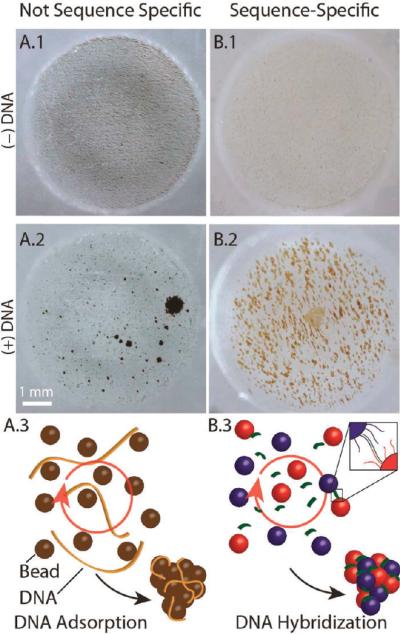
Figure 1.
Chaotrope-driven aggregation (CDA) forms “pinwheels” in a microfluidic chip well. (A) Magnetic silica beads (8 μm) in 20 μL of 6 M GdnHCl in a RMF. (A.1) In the absence of DNA, the beads are rapidly (20 s) distributed throughout the microwell. (A.2) Adding 1 ng of hgDNA and exposure to a RMF for 60 s causes the beads to aggregate into pinwheels that are easily detectable with the naked eye. (A.3) Schematic of a possible mechanism for CDA. (B) Superparamagnetic beads (1 μm) with adducted oligonucleotides specific to the 3′ and 5′ sequences of a DNA target under nonchaotropic conditions. (B.1) In the absence of target DNA, the beads are distributed throughout the microwell. (B.2) Response of beads to the addition of the DNA target in a RMF. (B.3) Schematic of a possible mechanism for hybridization-induced aggregation.