Abstract
The basic Helix-Loop-Helix (bHLH) domain is an essential highly conserved DNA-binding domain found in many transcription factors in all eukaryotic organisms. The bHLH domain has been well studied in the Animal and Plant Kingdoms but has yet to be characterized within Fungi. Herein, we obtained and evaluated the phylogenetic relationship of 490 fungal-specific bHLH containing proteins from 55 whole genome projects composed of 49 Ascomycota and 6 Basidiomycota organisms. We identified 12 major groupings within Fungi (F1–F12); identifying conserved motifs and functions specific to each group. Several classification models were built to distinguish the 12 groups and elucidate the most discerning sites in the domain. Performance testing on these models, for correct group classification, resulted in a maximum sensitivity and specificity of 98.5% and 99.8%, respectively. We identified 12 highly discerning sites and incorporated those into a set of rules (simplified model) to classify sequences into the correct group. Conservation of amino acid sites and phylogenetic analyses established that like plant bHLH proteins, fungal bHLH–containing proteins are most closely related to animal Group B. The models used in these analyses were incorporated into a software package, the source code for which is available at www.fungalgenomics.ncsu.edu.
Keywords: bHLH, fungal, phylogeny, discriminant, analysis
Introduction
The basic Helix-Loop-Helix (bHLH) domain is a highly conserved DNA-binding motif found in Eukarya and Bacteria that is involved in a number of important cellular signaling processes; including differentiation, metabolism, and environmental response (Robinson and Lopes 2000; Jones 2004; Castillon et al. 2007). Proteins containing the bHLH domain compose a superfamily of transcription factors commonly found in large numbers within plant, animal, and fungal genomes (Murre et al. 1989; Riechmann et al. 2000; Ledent and Vervoort 2001). Across such transcription factors, the bHLH domain is evolutionarily conserved while little sequence similarity exists beyond the motif itself (Carretero-Paulet et al. 2010).
The ∼60 amino acid bHLH region is divided into two main components: basic and dimerization regions. The first 13 N-terminal amino acids are responsible for DNA interaction; generally containing 5 to 6 basic residues that facilitate DNA binding (Massari and Murre 2000). Many bHLH domains bind to the hexanucleotide sequence known as the E-box (CANNTG). The dimerization region consists of two amphipathic alpha-helices separated by a loop of variable length. These alpha-helices either homodimerize or heterodimerize to a secondary alpha-helix containing protein to facilitate transcription (Ma et al. 1994; Shimizu et al. 1997).
The bHLH domain was first elucidated in Animals where bHLH proteins have been grouped into six major groups (A–F) based on evolutionary relatedness, DNA-binding motifs, and functional properties (Atchley and Fitch 1997; Ledent and Vervoort 2001). Group A includes proteins such as MyoD, dHand, Twist, and E12. Group A sequences bind the E-box sequence CAGCTG or CACCTG and are identified by containing an R at position 8 in the basic region. Group B sequences are known to bind the E-box sequence CACGTG, containing a histidine (H) or lysine (K) at position 5 and an arginine (R) at position 13 in the basic region. Members of Group B include Myc, Mad, Max, SREBP and Tfe. Many Group B proteins are known to contain an additional leucine zipper domain directly adjacent to the second helix. Group C members, such as Sim, Trh, and Ahr, have a conserved downstream Per-Arnt-Sim (PAS) domain that facilitates dimerization to other PAS-containing proteins and generally bind non-E-box sequences. Group D includes Id and Emc, however they lack a conserved basic region and act as transcription regulators through heterodimerization (Fairman et al. 1993). Group E proteins bind the target sequence CACGNG, contain a proline (P) in the basic region at site 6, and consist of members such as E(spl), Gridlock, Hairy, and Hey. Finally, Group F consists of COE-bHLH proteins, having more divergent bHLH sequences when compared with Groups A–E and containing an additional PAS domain (Pires and Dolan 2009).
Early studies of plant bHLH proteins primarily focused on Arabidopsis thaliana and Oryza sativa, which contain 167 and 177 bHLH sequences, respectively, compared with 39 and 125 in Caenorhabditis elegans and Homo sapiens, respectively (Ledent et al. 2002; Carretero-Paulet et al. 2010). With the recent abundance of genome initiatives, current studies include a more diverse selection such as algae, bryophytes, and other land plants. In contrast to animals, phylogenetic analyses of plant bHLH proteins classify them into 26–33 subgroups (Buck and Atchley 2003; Pires and Dolan 2009; Carretero-Paulet et al. 2010). Characterized members within these groups influence many biological processes including light and hormone signaling (Ni et al. 1998; Friedrichsen et al. 2002), wound and drought response (Smolen et al. 2002), fruit and flower development (Liljegren et al. 2004; Szécsi et al. 2006), and stomata and root development (Menand et al. 2007; Pillitteri et al. 2007).
Phylogenetic analyses suggest that plant sequences are most related to animal Group B (Buck and Atchley 2003; Heim et al. 2003). From the few fungi included in these studies, it has been noted that fungal sequences also appear to share most similarity to Group B (Atchley and Fitch 1997; Ledent and Vervoort 2001; Atchley and Fernandes 2005).
Here, we conduct a comprehensive analysis of bHLH-containing proteins from 55 completed fungal genomes encompassing Ascomycota and Basidiomycota organisms. Classification of these proteins is essential for understanding the evolutionary diversification of the bHLH domain and the biological roles they play in fungal organisms. Using a variety of bioinformatic and phylogenetic tools, we were able to identify and characterize 12 conserved bHLH fungal groups and determine patterns of gain and loss of bHLH proteins from a taxonomic perspective. Several statistical tools were then applied to evaluate the fundamental molecular architecture differences between the 12 fungal groups, including several classification models to accurately distinguish sequences into the groups. Some models not only distinguished groups but also provided a measure of the biological significance of discerning amino acid sites. These models were then tested against a larger set of known bHLH sequences, providing a measurement for the performance of each model. Finally, we show that, like plants, fungal bHLH are most closely related to animal Group B, suggesting that animal Groups A, C–E were likely not present in the metazoan common ancestor. The models, sequence data, and source code obtained and built for these analyses were incorporated into a software package available at www.fungalgenomics.ncsu.edu.
Materials and Methods
Whole Genome Fungal bHLH Sequence Identification and Analysis
Fungal bHLH sequences were aligned against plant and animal bHLH amino acid sets available from previous work (Atchley et al. 2000; Atchley and Zhao 2007). Each fungal sequence was aligned to these expert sets using an iterative approach that retained the length and structure of the bHLH domain as follows (Ferré-D’Amaré et al. 1993; Atchley et al. 1999). 1) A full-length protein sequence was chosen from the set to be aligned. 2) BLAST (Altschul et al. 1997) was used to identify up to ten orthologs from the expert sets, choosing hits with the lowest e-value. 3) The query and orthologs were then globally aligned with MUSCLE 5.0 (Edgar 2004). 4) The alignment was then evaluated for retention of the bHLH structure; that is, there were no gaps inserted into either the query or orthologs within the basic, Helix 1 or Helix 2 subdomains. 5) The newly aligned bHLH motifs contained in the query sequences were then placed into the expertly aligned set or the query sequence was placed back into the unaligned set depending on fulfillment of step 4. Steps 1–5 were repeated until most query sequences were aligned. Those few sequences still not aligned were then manually edited to meet bHLH domain requirements. This resulted in a new sequence data set of expertly aligned fungal bHLH domains.
Consensus sequences were determined by using the “50-10” rule (Carretero-Paulet et al. 2010). A given site of the bHLH domain was included in the consensus sequence if an amino acid at that site was present in over 50% of the sequences. For each site included in the consensus, an additional amino acid was added if it existed in at least 10% of all sequences.
The Boltzmann–Shannon entropy value was calculated for each site in the sequence alignment for fungal sequences. To determine the normalized group entropy value: 1) amino acids were grouped based on molecular characteristics (acidic, basic, aromatic, aliphatic, aminic, hydroxylated, cysteine, and proline) resulting in eight sets (DE, HKR, FWY, AGILMV, NQ, ST, C, and P, respectively) (Atchley et al. 1999; Wang and Atchley 2006); 2) The Boltzmann–Shannon entropy values, based on individual amino acids and the eight amino acid groups, were calculated at each site (Atchley and Fernandes 2005); 3) The entropy values were normalized to range from 0 to 1, with respect to possible minimum and maximum values, respectively. Amino acid sites were then interpreted from conserved to variable based on entropy values closer to 0 or 1, respectively.
Conserved motifs within bHLH-containing proteins were identified using MEME 3.5.7 (Bailey and Elkan 1994). Meme parameters: minimum motif width, 8; maximum motif width, 100; and maximum motifs to find, 50. Functionality of detected motifs was determined, where possible, by evaluating said motifs through MAST (Bailey and Gribskov 1998), NCBI’s Conserved Domain Database (Marchler-Bauer et al. 2009), Prosite (Sigrist et al. 2010), and InterPro (Hunter et al. 2009).
Phylogenetic Analysis by Taxonomic Grouping
Evolutionary relationships of the bHLH domain were determined in the same manner for several different fungal sequence data sets (all fungi; Basidiomycota; Pezizomycotina; and Saccharomycotina). Each data sets’ phylogeny was determined with maximum likelihood (ML), neighbor-joining (NJ), and maximum parsimony (MP) analyses. Bayesian analysis (BA) was conducted on the entire set of plant, animal, and fungal-aligned bHLH domain sequences.
ProtTest 1.4 (Abascal et al. 2005) was used to determine the best fit amino acid substitution model and parameter values for each data set. In each case, the Le and Gascuel (2008) (LG) model with an estimated γ-distribution parameter (G) and the proportion of invariant sites (I) was the best fit according to the Akaike information criterion; with the “JTT + I + G” (Jones, Taylor, Thorton) model a close second.
PHYML 2.4.5 (Guindon and Gascuel 2003), with the “LG + I + G” model, was used to run the ML analysis. The invariant sites and γ-parameter were set to values obtained with ProtTest and eight relative substation rate categories to correct for the heterogeneity of amino acid substitution rates. The Subtree Pruning and Regrafting method was used to search tree topology. Branch support for the resulting topology was determined by both the Shimodaria–Hasegawa-like approximate likelihood ratio test and a 1,000 replicate bootstrap analysis.
MEGA 4.0 (Tamura et al. 2007) was used to run the NJ and MP analyses, including a 1,000 replicate bootstrap test to estimate topology support. The JTT + I + G model was used for the NJ analysis. The NJ running options used were: 1) Pairwise deletion for Gaps/Missing data to account for highly variable sites, specifically in the loop subdomain; 2) rates among sites was set to Gamma distributed; 3) the value for the γ-value determined by ProtTest for the Gamma parameter. For the MP analysis, the Gaps/Missing data parameter was set to “Use All Sites” to account for variable amino acid sites.
BA was performed with MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003) with the following parameters: two independent runs with four Markov chains each, 10 million generations, sampling every 1000th generation, invgamma model, and eight categories. The standard deviation of split frequencies was below 0.01 at generation 10 millions, at which point a consensus tree was constructed from 1,800 trees (900 from both runs) after first discarding 100,000 generations as burn-in.
Classification Models
Decision trees (Breiman et al. 1984; Atchley and Zhao 2007) were built using SAS software, Enterprise Miner 5.2. A chi-square test with a significance level of 20% was used as the splitting criteria. The bifurcating tree was limited to a depth of 5 nodes, requiring a minimum of ten observations for a split and at least four observations per leaf.
Following the data transformation process described in Atchley and Zhao (2007), amino acids for each sequence were transformed into a 1 × 5 vector of factor scores using the HDMD package (McFerrin 2010). Factor scores are quantitative values for amino acids based on amino acid properties. The five-factor scores, which can be interpreted as independent physiochemical indices, were derived by Atchley and Fernandes (2005), from 495 measurable amino acid properties. The factor scores (pah; pss; ms; cc; and ec) are associated with biological properties (polarity, accessibility, and hydrophobicity; propensity for secondary structure; molecular size or volume; codon composition; and electrostatic charge; respectively). Factor scores are independent, thus, we created an additional data set containing the combination of all five-factor scores (all). This resulted in the total of six-factor score transformed data sets: pah, pss, ms, cc, ec, and all from the 488 grouped fungal sequences.
Discriminant analyses (Johnson and Wichern 2001), canonical variate analysis (CVA), and stepwise discriminant analysis (SWDA) were used to build models on all six-factor score data sets to evaluate molecular differences between the 12 fungal group sequences. These discriminant analyses were used to define the latent structure of covariation among-groups and obtain a set of amino acid sites that best differentiate between the groups in the fungal group data sets.
The step-up SWDA procedure was used to rank amino acid sites based on their ability to discriminate defined groups (r2) (Atchley and Zhao 2007). In the step-up procedure, variables (amino acid sites) were added sequentially (step) based on the site’s discriminating power. Amino acid sites were added until an average squared canonical correlation (ASCC) reached a value of 70% for pah, pss, ms, cc, ec and 80% for all data sets. The ASCC describes the related distinctiveness of the groups at a given step in the model, meaning a 100% ASCC would imply complete discrimination between the defined groups. SAS software, Version 9.2, was utilized in the SWDA. Those variables with r2 > 70% were considered the most discerning sites.
CVA assesses the discriminatory ability of all variables (factor score transformed amino acid sites) simultaneously to generate a linear model to differentiate between defined groups. The CVA includes the calculation of eigenvectors (canonical variates [CVs]) from the among-group covariance matrix. CVA for the six-factor score data sets resulted in 11 CVs for each analysis. The square root of the Mahalanobis pairwise distance was also calculated, providing a relative measure of the divergence between groups. CVA and plotting of CVs were conducted utilizing the statistical software package R (R Development Core Team 2009). Amino acid sites were considered discerning if they met the following criteria: 1) contained within CVs that explained >5% of the among-group covariation; 2) had absolute magnitudes > 1 for the pah, pss, ms, cc, and ec analyses; and 3) had absolute magnitude > 8 for the all analysis.
Testing Methods
Fungal protein sequences annotated with a bHLH domain (707) were obtained from Interpro 31.0 (Hunter et al. 2009). A data set was then constructed for the testing of classification models from the 198 fungal sequences not used in model construction (F.198). These sequences were assigned fungal groups by utilizing BLAST to find homologous sequences that had a priori defined groups. In the few instances where a sequence aligned to more than one fungal group, assignation was based on majority rule. The bHLH sequences in F.198 were evaluated as follows: 1) the bHLH domain was extracted from the full amino acid sequence; 2) transformed into factor scores; 3) subjected to several classification methods as described under classification methods. F.198 was used to test the performance of each classification method.
To determine the performance of the classification models, confusion matrices were generated by classifying sequences from the F.198 data set. We then measured the sensitivity (ability to identify positive results) and specificity (ability to identify negative results) for each model (data not shown). These measures were calculated from the “One versus All” approach commonly used with multiclass classification models (Rifkin and Klautau 2004). Finally, model performance was measured by determining the overall accuracy (ability to correctly identify results) and its assessment (coefficient of agreement; eq. 1) (Gross 1986; Tsoumakas and Katakis 2007). Good accuracies have assessments >80%.
| (1) |
where for a given confusion matrix: N = number of trials, k = number of states (rows and columns), xii = value at row i and column i of matrix.
Results
Identification of Fungal bHLH Sequences from Whole Genome Projects
Previous bHLH analyses have focused primarily on animal or plant sequences, with token references to fungal organisms (Buck and Atchley 2003; Heim et al. 2003; Atchley and Fernandes 2005; Li et al. 2006). This has provided insightful, but limited, information of the phylogenetic relationship of fungal bHLH sequences to those of plants and animals. Nevertheless, it has provided no insight into the diversity of the bHLH domain within Fungi.
To obtain fungal-specific gene sequences containing the bHLH domain, we utilized the protein sequence analysis and classification database InterPro. Using protein signatures built on known bHLH domains (IPR001092) and classified based on taxonomy, we identified 707 fungal bHLH–containing sequences. From this set, 198 sequences not belonging to whole genome projects or originating from projects with incomplete assemblies and gene calls were set aside. This resulted in 509 full amino acid sequences putatively containing the bHLH domain from 55 genome projects representing major evolutionary fungal lineages, encompassing the Ascomycota (49 members) and Basidiomycota (6 members) Phylums. An iterative global alignment to a reference set of 147 plant bHLH (Buck and Atchley 2003) and 284 animal bHLH domains (Atchley and Zhao 2007) resulted in the identification and alignment of all 509 fungal bHLH domains (supplementary data 1, Supplementary Material online). The location of each bHLH domain in each protein sequence predicted by the protein signature from InterPro directly corresponded to the location of the domain determined through our alignment method (supplementary data 1, Supplementary Material online). Using this iterative global alignment approach, we were able to ensure direct comparison of homologous amino acids by enumerating the bHLH domain as described in previous work on Animals (region: basic, first Helix, Loop, second Helix; sites: 1–13, 14–28, 29–49, 50–64; respectively) (Atchley and Fitch 1997).
We identified 34 perfect duplicate bHLH domains within eight fungal species (data not shown). Duplicate bHLH domains arose for a variety of genome sequencing artifacts (including inconsistent gene calls and strain-specific sequencing differences) and were not likely due to recent sequence duplications. A representative was chosen from each set of duplicates, resulting in 19 sequences being removed from our analyses. The remaining 490 bHLH fungal sequences from 422 Ascomycota and 59 Basidiomycota proteins are shown in table 1 arranged by organism and taxonomy.
Table 1.
Gene Count Summary of the 12 Fungal bHLH Groups by Completed Fungal Genome.
| Taxonomy | F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | F11 | F12 | Un | Tot |
| Basidiomycota | ||||||||||||||
| Ustilaginomycotina | ||||||||||||||
| Malassezia globosa | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||||
| Ustilago maydis | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 12 | |||
| Agaricomycotina | ||||||||||||||
| Postia placenta | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||||
| Laccaria bicolor | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 14 | |||
| Coprinopsis cinerea | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 10 | ||||
| Filobasidiella neoformans | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 9 | |||||
| Ascomycota | ||||||||||||||
| Taphrinomycotina | ||||||||||||||
| Schizosaccharomyces japonicus | 1 | 2 | 1 | 4 | ||||||||||
| Schizosaccharomyces pombe | 1 | 2 | 1 | 4 | ||||||||||
| Saccharomycotina | ||||||||||||||
| Saccharomycetes, Saccharomycetales | ||||||||||||||
| Metschnikowiaceae | ||||||||||||||
| Clavispora lusitaniae | 2 | 1 | 3 | 1 | 1 | 8 | ||||||||
| Dipodascaceae | ||||||||||||||
| Yarrowia lipolytica | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 9 | |||||
| Candida (mitosporic Saccharomycetales) | ||||||||||||||
| Candida dubliniensis | 2 | 1 | 3 | 1 | 1 | 1 | 9 | |||||||
| Candida tropicalis | 2 | 3 | 1 | 1 | 7 | |||||||||
| Candida albicans | 2 | 1 | 3 | 1 | 1 | 8 | ||||||||
| Saccharomycetaceae | ||||||||||||||
| Pichia pastoris | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 10 | |||||
| Lachancea thermotolerans | 2 | 1 | 2 | 1 | 1 | 1 | 8 | |||||||
| Vanderwaltozyma polyspora | 2 | 1 | 3 | 1 | 1 | 8 | ||||||||
| Eremothecium gossypii | 2 | 1 | 2 | 1 | 1 | 7 | ||||||||
| Kluyveromyces lactis | 2 | 1 | 3 | 1 | 1 | 8 | ||||||||
| Candida glabrata | 2 | 1 | 3 | 1 | 1 | 1 | 9 | |||||||
| Zygosaccharomyces rouxii | 2 | 1 | 2 | 1 | 1 | 1 | 8 | |||||||
| Saccharomyces cerevisiae | 2 | 1 | 2 | 1 | 1 | 1 | 8 | |||||||
| Debaryomycetaceae | ||||||||||||||
| Lodderomyces elongisporus | 2 | 1 | 2 | 1 | 1 | 7 | ||||||||
| Debaryomyces hansenii | 2 | 1 | 2 | 1 | 1 | 1 | 8 | |||||||
| Meyerozyma guilliermondii | 2 | 1 | 3 | 1 | 1 | 8 | ||||||||
| Scheffersomyces stipitis | 2 | 1 | 2 | 1 | 1 | 7 | ||||||||
| Ascomycota | ||||||||||||||
| Pezizomycotina | ||||||||||||||
| Dothideomycetes | ||||||||||||||
| Pyrenophora tritici-repentis | 1 | 1 | 4 | 2 | 1 | 1 | 1 | 11 | ||||||
| Phaeosphaeria nodorum | 1 | 1 | 4 | 1 | 2 | 1 | 1 | 1 | 12 | |||||
| Leotiomycetes | ||||||||||||||
| Sclerotinia sclerotiorum | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 9 | |||||
| Botryotinia fuckeliana | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 9 | |||||
| Sordariomycetes | ||||||||||||||
| Nectria haematococca | 1 | 1 | 5 | 1 | 1 | 1 | 1 | 1 | 12 | |||||
| Magnaporthe oryzae | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 10 | |||||
| Chaetomium globosum | 1 | 4 | 1 | 2 | 1 | 1 | 1 | 11 | ||||||
| Podospora anserina | 1 | 1 | 8 | 1 | 2 | 1 | 1 | 1 | 16 | |||||
| Sordaria macrospora | 1 | 1 | 6 | 1 | 1 | 1 | 1 | 12 | ||||||
| Neurospora crassa | 1 | 1 | 7 | 1 | 1 | 1 | 1 | 1 | 14 | |||||
| Eurotiomycetes | ||||||||||||||
| Onygenales | ||||||||||||||
| Trichophyton verrucosum | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||||
| Arthroderma benhamiae | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||||
| Arthroderma otae | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||||
| Ajellomyces dermatitidis | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||||
| Ajellomyces capsulatus | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |||||||
| Uncinocarpus reesii | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||||
| Paracoccidioides brasiliensis | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | ||||||
| Coccidioides posadasii | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | |||||
| Eurotiales | ||||||||||||||
| Talaromyces stipitatus | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 10 | |||||
| Emericella nidulans | 1 | 4 | 1 | 2 | 1 | 1 | 1 | 1 | 12 | |||||
| Neosartorya fischeri | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 10 | |||||
| Aspergillus fumigatus | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 9 | ||||||
| Penicillium chrysogenum | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 10 | |||||
| Penicillium marneffei | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 10 | |||||
| Aspergillus niger | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 9 | |||||
| Aspergillus terreus | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 10 | |||||
| Aspergillus flavus | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 11 | |||||
| Aspergillus oryzae | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 10 | ||||||
| Aspergillus clavatus | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 9 |
Note.—Listed fungal organisms have completed genome projects, fully annotated gene sets, and contain bHLH genes. A simplified taxonomic classification, the total bHLH copy count, and the bHLH copy count within fungal groups F1–F12 are provided for each organism.
The number of bHLH proteins in the fungal genomes ranged from a maximum of 16 (Podospora anserina) to as few as four within the Taphrinomycotina Subphylum (table 1). Members of Saccharomycotina Subphylum typically contained eight bHLH sequences; however some contained nine or ten proteins while Candida tropicalis, Eremothecium gossypii, Lodderomyces elongisporus, and Scheffersomyces stipitis each contained only seven. The Sordariomycetes class members contained between 10 and 16 members with a median of 12, whereas members of the Eurotiomycetes class ranged between 7 and 11. The Onygenales and Eurotiales orders, within the Eurotiomycetes class, typically contained eight and ten proteins each, respectively. The number of bHLH proteins in Basidiomycota ranged from 7 to 14. An insufficient number of sequenced taxa were available to identify clear patterns within the Basidiomycota Phylum. In summary, we observed distinct differences in the typical number of bHLH proteins within the Sordariomycetes and Saccharomycetes classes and the Onygenales and Eurotiales orders.
Positional Conservation and Consensus Motif
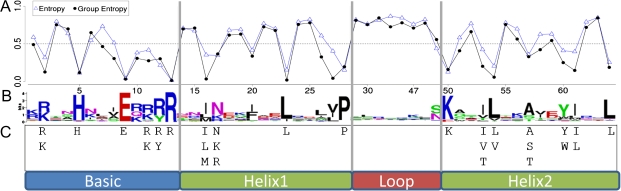
To determine the conservation of amino acid sites of the fungal bHLH domain, we performed Boltzmann–Shannon entropy and group entropy analyses (Atchley et al. 1999, 2000; Wollenberg and Atchley 2000), generated a bit score weblogo (Crooks et al. 2004), and determined the consensus sequence motif (fig. 1) on the set of nonredundant aligned sequences.
FIG. 1.
Fungi bHLH entropies, logos, and consensus. (A) The bHLH normalized group entropy by position. Lower values indicate conservation, whereas values close to one approach complete randomness. (B) The graphical representation of the amino acids at each position of the bHLH domain. Symbols representing amino acids are scaled by their bit score (a derivation of entropy) at a given position. (C) The 50-10 consensus sequences for fungi. Using an alignment of bHLH domains, amino acids occurring at a frequency of more than 50% at a given site are displayed. At each of these sites, additional amino acids are displayed beneath if they are conserved in 10% or more of the sequences.
We evaluated the conformity of bHLH sequences to the entire fungal set by determining the number of mismatches between each sequence and the consensus sequence (supplementary data 1, Supplementary Material online). In previous work, sequences were considered highly divergent and removed from subsequent analyses if they contained more than eight to ten mismatches to the consensus sequence (Buck and Atchley 2003; Heim et al. 2003; Toledo-Ortiz et al. 2003). We retained all 490 fungal bHLH sequences, as there were no sequences with more than seven such mismatches.
We identified 17 conserved positions in fungi based on amino acid frequency (table 2). Six additional conserved sites were identified based on low-group entropy (conserved amino acid properties). As shown in figure 1, in the basic region (sites 1–13) of the fungal consensus motif, amino acid positions 2, 5, 9, and 13 had low entropies, high bit scores, and were represented by amino acids R, H, E, and R, respectively, at a frequency of at least 50%. Sites 8, 10, 11, and 12 were considered moderately conserved, having group entropies between 0.276 and 0.308. Sites 16, 23, and 28 were highly conserved in the first Helix (sites 14–28), having I, L, and P amino acids at frequencies of 59%, 88%, and 92%, respectively. Site 27 had high entropy but low-group entropy being highly conserved for aliphatic amino acids, with V, I, L, and M at frequencies of 49%, 24%, 21%, and 3%, respectively. Additionally, moderately conserved Helix 1 sites 17, 20, and 26 had group entropies between 0.327 and 0.366. In Helix 2 (sites 50–64), highly conserved sites included 50, 53, 54, 60, 61, and 64. Each of these sites had amino acids K, I, L, Y, I, and L at frequencies of 90%, 58%, 84%, 68%, 67%, and 85% respectively. Site 57 contained A in over 50% of fungal sequences, however, could only be considered moderately conserved as it had entropy and group entropy values of 0.357 and 0.330, respectively.
Table 2.
Structural Attributes and Significant Sites of the bHLH Domain.
| Site | Structural | CS | DT | SM | pah | pss | ms | cc | ec | all |
| 1 | DP | √ | √ | C | ||||||
| 2 | DP | √ | C | C | ||||||
| 3 | ||||||||||
| 4 | √ | √ | ||||||||
| 5 | DP | √ | C | C | ||||||
| 6 | P | √ | SC | S | C | C | SC | SC | ||
| 7 | √ | √ | ||||||||
| 8 | DP | * | √ | √ | SC | SC | C | SC | ||
| 9 | DP | √ | C | C | C | |||||
| 10 | P | * | C | C | SC | S | C | |||
| 11 | P | * | √ | S | SC | C | ||||
| 12 | DP | * | √ | √ | SC | SC | SC | SC | SC | SC |
| 13 | DP | √ | C | C | C | C | C | |||
| 14 | S | |||||||||
| 15 | P | √ | √ | SC | SC | |||||
| 16 | B | √ | √ | C | C | C | ||||
| 17 | P | * | C | C | C | C | ||||
| 18 | ||||||||||
| 19 | √ | √ | S | S | ||||||
| 20 | B | * | √ | √ | C | C | C | C | C | |
| 21 | ||||||||||
| 22 | ||||||||||
| 23 | B | √ | C | C | C | C | C | |||
| 24 | S | |||||||||
| 25 | ||||||||||
| 26 | * | |||||||||
| 27 | B | √ | C | |||||||
| 28 | B | √ | ||||||||
| 50 | DPB | √ | √ | √ | C | SC | SC | C | SC | SC |
| 51 | S | SC | C | C | ||||||
| 52 | ||||||||||
| 53 | B | √ | √ | C | C | |||||
| 54 | B | √ | C | SC | C | |||||
| 55 | ||||||||||
| 56 | ||||||||||
| 57 | * | |||||||||
| 58 | ||||||||||
| 59 | ||||||||||
| 60 | √ | |||||||||
| 61 | B | √ | ||||||||
| 62 | ||||||||||
| 63 | ||||||||||
| 64 | B | √ |
Note.—The molecular architecture of bHLH positions is compiled from previous work on crystalline structures of animal proteins. Structural attributes noted are DNA contact of the E-box (D), phosphate backbone contact (P), or buried site within the hydrophobic core of the dimerized helices (B). Highly (√) and moderately (*) conserved sites are denoted (CS). Sites integral in the decision tree analysis (DT) and the simplified model (SM) are also reported. Last, SWDA (S) and CVA (C) significant sites are shown within each factor score data set (pah, pss, ms, cc, ec, and all).
A number of these sites are conserved in plant and animal bHLH domains (Ferré-D’Amaré et al. 1993). At site 9, glutamic acid (E) was present in over 90% of E-box binding animal proteins and has been shown to directly contact DNA (Atchley et al. 1999; Pires and Dolan 2009). In a recent plant study, site 9 was represented by E in more than 74% of such sequences (Pires and Dolan 2009). We found that in Fungi, >98% of bHLH sequences contained an E at site 9.
Site 28 is another highly conserved site that has a conserved P that breaks the first Helix and starts the loop region. This highly conserved site in Plants and Animals contained P in 92% of fungal sequences. Sites 23 and 64 contained L (helix stabilization) in over 80% of plant and animal sequences (Pires and Dolan 2009) and over 85% of fungal sequences. Aliphatic amino acids, essential for dimerization, were conserved within sites 54 and 61 at 98% and 89% in Fungi and over 98% and 93% in Animals and Plants. The presence of these highly conserved sites demonstrates that the fungal bHLH domain shares similar architecture to those identified in Plants and Animals.
Phylogenic Analysis of Fungal Sequences
To elucidate the evolutionary relationships between bHLH domains within and between fungal lineages, we determined the phylogeny of sequences in four data sets (Basidiomycota, Pezizomycotina: filamentous members of Ascomycota, Saccharyomycotina: yeast-like members of Ascomycota, and all Fungi) using five phylogenetic analyses (ML, NJ, MP, ML Bootstrap, and BA). Based on high support values, tree topology, branch lengths, and majority support from each phylogeny, the 59 Basidiomycota, 286 Pezizomycotina, and 137 Saccharyomycotina bHLH proteins were split into 11, 9, and 10 clades, respectively (supplementary fig. S1A–C, Supplementary Material online). Based on the same methods, the 490 fungal bHLH proteins were split into 12 major clades (fungal groups F1–F12) (supplementary fig. S1D, Supplementary Material online). Annotated sequences, where available, shared similar biological and molecular functions with their group members (table 3). Each group was further supported by conserved loop length (Buck and Atchley 2003), consistency of basic amino acids in the basic subdomain (Atchley and Fitch 1997), and low divergence from the consensus sequence (supplementary data 1, Supplementary Material online). Several groups had average loop lengths of >40 amino acids, uncommon in either plant or animal bHLH sequences (supplementary data 1, Supplementary Material online). However, sequences with these extended loops typically were found in the same clade, such as F2. Conservation of such clades across Basidiomycota and Ascomycota fungi possibly arose from additional functionality provided by an elongated loop.
Table 3.
Known Biological and Molecular Functions for bHLH Proteins by Fungal Group.
| Group | Reported Members | Biological Function |
| F2 | RTG1, RTG3, MGG_05709 | Interorganelle communication between mitochondria, peroxisomes, and nucleus. |
| F3 | CBF1, CBF1P, CaCBF1, AnBH1, CPF1 | Chromosome segregation, methionine auxotrohpic growth, rRNA transcription, repression of penicillin biosynthesis, regulation of sulfur utilization, ribosome biogenesis, and glycolysis. |
| F4 | TYE7, SAH-2, HMS1, SRE1, SRE2, CPH2, CAP1P | Sexual development, aerial hyphae development, hypoxic response, carbon catabolite transcription activation, regulation of glycolysis, ergosterol biosynthesis, heme biosynthesis, phospholipid biosynthesis. |
| F5 | Q6MYV5 | Nitrate assimilation, quinate utilization. |
| F6 | PHO4, NUC-1, PalcA | Response to copper ion, regulate phosphate acquisition and metabolic process, promotes sexual development, represses asexual development. |
| F8 | ESC1, devR | Sexual differentiation, sexual conjugation, development under standard growth conditions. |
| F10 | YAS2 | Alkane response. |
| F11 | INO4, YAS1 | Derepression of phospholipid synthesis, alkane response. |
| F12 | INO2 | Derepression of phospholipid synthesis. |
Note.—No functional annotations were found for members of groups F1 and F7. Literature describing biological functions of the reported members of groups F2–F12 are cited in the manuscript.
Each fungal group was composed of one or more clades from the Basidiomycota (B1–B12), Pezizomycotina (P1–P12), or Saccharyomycotina (S1–S12) phylogenies, as denoted on the ML tree in figure 2. Groups B1–B12, P1–P12, and S1–S12 were enumerated to reflect their associated fungal group, for example, B1 is a clade within F1. Based on the composition of each fungal group, many bHLH domain gains and losses have occurred since the most recent common ancestor (MRCA) between Basidiomycota, Pezizomycotina and Saccharomycotina organisms. The MRCA likely contained bHLH domains found in F2–F5 and F11 as Basidiomycota, Pezizomycotina, and Saccharomycotina organisms were all represented in these groups. Additionally, we observed expansion of F2 but not F3 in the Saccharomycotina subphylum (table 1). Basidiomycota and Pezizomycotina fungi were represented in groups F8 and F10 but lost from the Saccharomycotina branch since the MRCA. Similarly, we observed that Pezizomycotina fungi have lost bHLH representation in F9 since the MRCA. F6 was either gained by the MRCA of Pezizomycotina and Saccharomycotina subphylums or was present in the MRCA and lost by Basidiomycotas. Finally, Saccharomycotina fungi have gained novel bHLH sequences present in F12 and Basidiomycota fungi in F1 and F7. Expansion and loss patterns were also observed at various taxonomic ranks within the Basidiomycota and Ascomycota Phylums (table 1). In F4, most fungi within the Ascomycota phylum experienced large expansions (2–8 copies), except for members of the Onygenales order (1 copy). Podospora anserina had the largest expansion in F4 sequences, accounting for half of its 16 bHLH sequences.
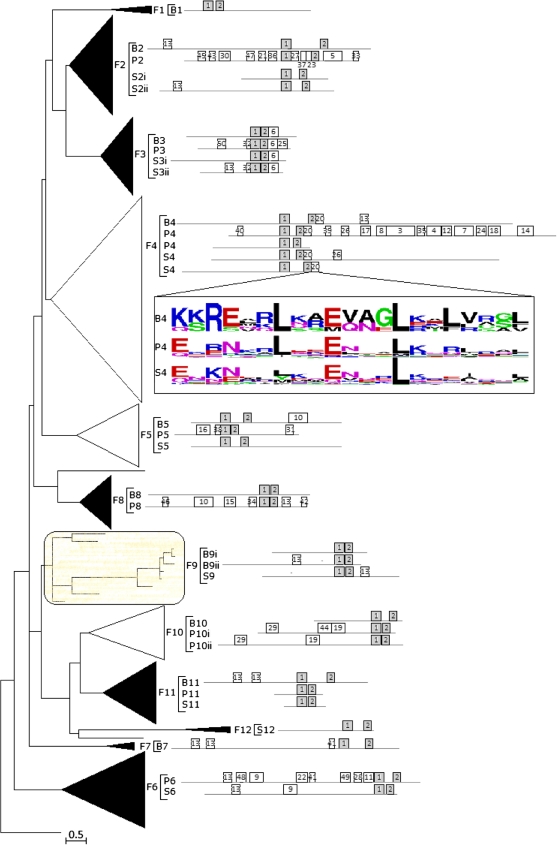
FIG. 2.
Phylogenetic analysis of fungal bHLH. Phylogenetic relationships, taxonomic representation, bHLH motif statistics, and architecture of conserved protein motifs for 12 fungal bHLH groups. ML tree of 490 fungal bHLH proteins (full representation of Basidiomycota, Pezizomycotina, Saccharyomycotina, and Fungal trees available in supplementary fig. S1A–D, Supplementary Material online). The tree is drawn to scale (branch lengths proportional to evolutionary distances) and has been rooted with a single representative from Chlamydomonas reinhardtii. Groups, determined by clades with strong support, are collapsed as triangles with width and depth proportional to the size and sequence divergence of each group, respectively. Groups supported by bootstrap values >30 in NJ or maximum parsimony (MP) analyses are colored black. The shaded group F9 was ambiguously retrieved in NJ, MP, or BA trees. Ungrouped genes are indicated as single lines, and the scale bar represents the estimated number of amino acid replacements per site. Basidiomycota (B), Pezizomycotina (P), and Saccharyomycotina (S) clades associated with each fungal group are noted in brackets. The architecture of conserved motifs, as determined through MEME, is graphically represented as boxes drawn to scale. Box enumeration corresponds to specific motifs found by MEME (supplementary fig. 2C, Supplementary Material online). Grey boxes represent motifs that match the bHLH domain. Last of all, the sequence logo for B4, P4, and S4 for 21 amino acids downstream of the bHLH domain are shown (motif 20).
Several other taxonomic groups experienced expansion, such as Dothideomycetes members in F6 (2 copies), whereas the other Ascomycotas retained only a single representative. Within Basidiomycota fungi, an expansion of F9 occurred in the Agaricomycotinas as compared with the Ustilaginomycotinas, in which only Ustilago maydis had a single F9 sequence. Thus, we observed many instances of expansion and loss among taxonomic ranks, except within F3, which has retained constant representation in all taxonomic groups (1 copy). In summary, the phylogenetic analysis shows that fungal bHLH proteins form 12 groups, each correlated with sequence characteristics, such as conserved loop length. Many of these groups remain distinct throughout fungal evolution despite the dramatic diversification of fungi.
Expansion of F4 in Sordariomycetes
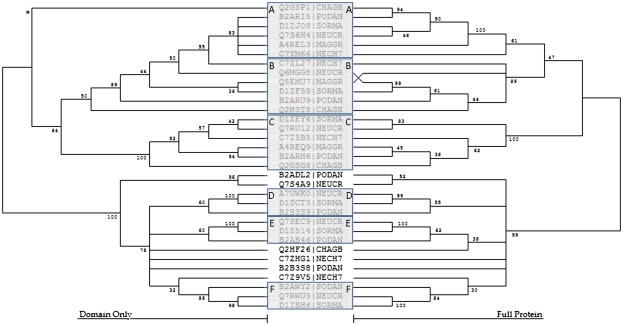
The most dramatic expansion observed was that of the Sodariomycetes within F4. Each member contained a minimum of three copies, with Sodaria macrospora, Neurospora crassa, and P. anserina having 6, 7, and 8 copies, respectively. To determine if the expansion was due to recent duplications within each organism or due to distinct bHLH sequences likely found in the Sodariomycete MRCA, we performed an additional phylogenetic analysis. Two NJ phylogenies were built, one from the bHLH domain and another from the entire bHLH-containing protein sequence (fig. 3). We found that the 33 sequences formed six distinct subclades each with bootstrap values of between 52 and 100 in both trees. Subclades A–C were composed of one copy from each Sodariomycete organism. Also, clades E–F each contained one protein from P. anserina, S. macrospora, and N. crassa. All members of subclade A were homologous to SRE1 and 2 (SREB) proteins. These findings support the MRCA containing an expansion of F4 rather than a large number of recent duplications in each of the Sodariomycete organisms. Additionally, these subclades generally support the published phylogeny of Sodariomycete organisms (Robbertse et al. 2006; Zhang et al. 2006; Nowrousian et al. 2010). With the notable exception of P. anserina, which shared six subclades with S. macrospora and N. crassa sequences but only three with Chaetomium globosum sequences.
FIG. 3.
NJ analyses of F4 proteins from Sordariomycetes organisms. The NJ trees were inferred from the bHLH domain only and from the entire bHLH-containing protein sequence. The trees are displayed with corresponding bootstrap values where branches with a bootstrap of less than 30 have been collapsed. Six strongly supported F4 subclades have been noted, where subclades A–C contain one member from each Sordariomycetes organism and Groups D–F each have one member from Podospora anserina, Sodaria macrospora, and Neurospora crassa. * The Q2GSP1 bHLH domain has been determined to be highly divergent by 1) containing seven mismatches to the fungal consensus motif, 2) possessing an uncharacteristic amino acid at site 12 for an F4 protein (D), and 3) containing a simple sequence repeat through both the basic and Helix 2 regions (DDDDDD). The full Q2GSP1 sequence, however, is strongly supported in the A subclade, suggesting the highly divergent bHLH arose from either evolutionary pressure or sequencing errors.
Conserved Motifs in Fungal bHLH Proteins
To identify conserved motifs in fungal bHLH proteins, we used MEME (Bailey and Elkan 1994) to search for 50 frequently occurring motifs in 490 sequences and correlated the results with Basidiomycota, Pezizomycotina, and Saccharomycotina groups (fig. 2). Motifs ranged in length from 11 to 86 amino acids were significant with e-values from 2.3 × 10−6014 to 2.7 × 10−146 and were nonoverlapping. The results provided additional support for Basidiomycota, Pezizomycotina, and Saccharomycotina group designations as the protein architecture (occurrence and location of motifs) was highly conserved within each fungal group.
The first and second most abundant motifs (motifs 1 and 2) corresponded to components of the bHLH domain noted as basic and Helix 1 regions and Helix 2 region, respectively. Both motifs were present in all sequences, with only a few exceptions. Pezizomycotina clade P2 was the only group to contain motifs that matched to the highly variable loop region where the average loop length was ∼63 amino acids. We also noted that the loop length between motifs 1 and 2 was exceptionally long in the Basidiomycota clade B2 with an average length of ∼70 amino acids. However, the B2 clade contained no identified motifs in the loop region. Therefore, the conserved domain within P2 loops may be an artifact of sampling of Pezizomycotina organisms or of the conserved nature of the full bHLH containing bHLH proteins within P2s. Thus, the loop remains a highly variable subdomain with undetermined function within Fungi.
Several motifs were found to be linked to functional properties besides the bHLH domain. For instance, motifs 3, 4, 7, 8, 12, 17, 26, and 35 were found in the C-terminal of many P4 proteins such as subclade A (figs. 2 and 3). These motifs were found to be part of ER membrane–bound transcription factors (sterol regulatory element-binding [SREB]). Within the fungal group F3, motif 6 was found to be related to functional components of the centromere-binding protein (CPB-1). Despite being found in many fungal bHLH sequences across several Basidiomycota, Pezizomycotina, and Saccharomycotina clades, the biological role of the highly repetitive motif 13 (Q-P-Q{22}) has yet to be defined.
The bHLH-ZIP domain consists of a conserved heptad leucine repeat (Leucine Zipper) adjacent to the bHLH domain. The bHLH-ZIP has been found in both plant and animal sequences, however, they are extremely divergent between Kingdoms with previous work supporting convergence (Atchley and Fitch 1997; Morgenstern and Atchley 1999; Pires and Dolan 2009). We found evidence of Leucine Zippers in fungal groups F2 and F4 and in Basidiomycota, Pezizomycotina, and Saccharomycotina clades B5, B7, P5, P10, and S11 (supplementary fig. S2A, Supplementary Material online). Motif 20, found extensively in F4, was composed of conserved leucines at downstream positions 7, 14, and 21 from the bHLH domain (fig. 2), indicative of the bHLH-ZIP domain. Motif 20 was the only motif with a known molecular function besides motifs 1 and 2 (bHLH). Although many motifs were linked to specific groups of bHLH–containing transcription factors, the role of these proteins and consequently the function of the majority of the motifs remain to be determined.
We observed that the spatial orientation of the bHLH domain with respect to the protein sequence (NH2-terminus, middle, or COOH terminus) was conserved within many of the Basidiomycota, Pezizomycotina, and Saccharomycotina groups (fig. 2). The approximate location of the bHLH domain within members of the fungal groups F3, F5, F8, and F10 was consistent within said groups. In addition, motifs 6, 9, 19, 20, 29, and 32 showed low spatial variation with respect to the bHLH domain. Conservation of special location within groups is likely indicative of a functional link between the motif, the bHLH domain, and the protein function.
Sequence Classification Using Decision Trees
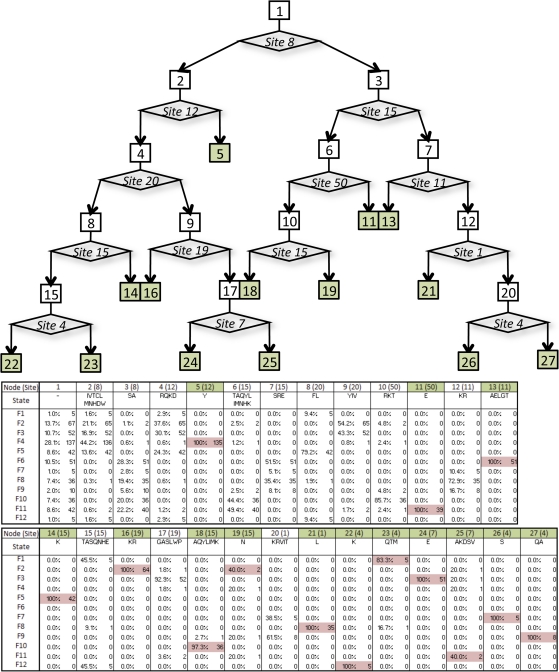
To identify key sites that distinguish fungal group sequences, we performed a decision tree analysis using the state of amino acid sites in the basic, Helix 1, and Helix 2 regions (fig. 4). Before starting the decision tree analysis, we created a new data set from the set of 490 fungal sequences by removing two sequences that were not placed into groups F1–F12. Starting with the entire data set of 488 sequences, each step bifurcates the data based on the amino acids at a given site. Steps are added until there are too few sequences to split, the tree hits a user set maximum depth, or the data subset converges on a group. Discerning sites 1, 4, 7, 8, 11, 12, 15, 19, 20, and 50 (table 2) accurately placed fungal sequences to their a priori defined groups with an accuracy rate for each group over 98% with the exception of F9 which was 88%. Overall, the accuracy of the decision tree was 95.5% (table 4).
FIG. 4.
Fungal decision tree analysis. The decision tree describing the separation of bHLH fungal groups F1–F12 by amino acid sites found in the bHLH domain. Each box of the figure represents a step in the decision tree which consist of a number of bHLH sequences from each fungal group and the amino acids at a given bHLH position (state). The sample size and proportion of group representatives is provided in the accompanying table. Diamonds contain the bHLH amino acid site that bifurcate the data into subsets of the previous state.
Table 4.
Validation of bHLH Classification Methods.
| Statistic | Decision Tree | CVA {pah} | CVA {pss} | CVA {ms} | CVA {cc} | CVA {ec} | CVA {all} | SWDA {all} | Simplified Model |
| 488 | |||||||||
| Accuracy | 95.5 | 100.0 | 99.6 | 99.6 | 98.4 | 99.8 | 100.0 | 99.2 | 99.0 |
| Coefficient | 94.7 | 100.0 | 99.5 | 99.5 | 98.1 | 99.8 | 100.0 | 99.1 | 98.8 |
| Unclassified | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 8 |
| 198 | |||||||||
| Accuracy | 92.9 | 96.1 | 96.1 | 97.2 | 93.9 | 96.1 | 95.0 | 95.4 | 96.4 |
| Coefficient | 92.0 | 95.6 | 95.6 | 96.8 | 93.0 | 95.6 | 94.3 | 94.8 | 95.9 |
| Unclassified | 0 | 19 | 19 | 19 | 19 | 19 | 19 | 3 | 3 |
Note.—The accuracy and coefficient of agreement are reported for the Decision Tree, SWDA{all}, each CVA, and the Simplified Model classification methods. The measures are derived from two data sets. The first measurements are based on the 488 fungal sequences used in building the models. The second set assesses the models on with the F.198 sequence set. The number of sequences that were unable to be classified (Unclassified) for each model are also reported.
All groups were accurately separated within 5 steps. For instance, all group F4 sequences were deduced in two steps: First, they contained an S or A at site 8 (step 2) and second, they had a Y at site 12 (step 5). The amino acid composition at discriminating sites used in the decision tree was readily visualized in the fungal group weblogos (supplementary fig. S2B, Supplementary Material online).
Sequence Classification Using Discriminant Analysis
To evaluate and compare the discriminating power each site had on separating groups F1–F12, we performed a stepwise discriminant analysis. Amino acid data for each site were transformed into numerical values by utilizing five numerical indices (factor scores) based on measured physiochemical amino acid properties as described in previous work (Atchley and Zhao 2007). Factor scores 1–5 have been linked to biological properties, including polarity, accessibility, and hydrophobicity (pah); propensity for secondary structure (pss); molecular size (ms); codon composition (cc); and electrostatic charge (ec), respectively. The transformed data set resulted in five numeric values for each amino acid for every position in each bHLH sequence (all).
To identify the discerning sites between fungal groups F1–F12, SWDA and CVA were performed on the factor score–transformed fungal data (Atchley and Zhao 2007) denoted as SWDA{factor score} and CVA{factor score}, respectively. SWDA pah, pss, ms, and cc each required more than 30 amino acid sites to explain >70% of the among group variance, where ec required only 20 sites (supplementary table S1, Supplementary Material online ). SWDA using the all data set, where each amino acid site was represented by five values, obtained an ASCC of 80% using 16 sites in 28 steps. These results showed that using only SWDA requires numerous amino acid sites to completely distinguish between the different fungal groups. However, SWDA did reveal a few highly discerning sites such as 6, 8, 12, 50, and 51.
The first CV of the CVA explained the vast majority of the variance in each of the six analyses, that is, revealed highly discerning sites (supplementary table S2, Supplementary Material online). For example, the pah, pss, ms, and cc CVA separated F4 from the other groups in the first CV. Additionally, the first CV in ec and all separated out group F11. Plotting the first and second CV of the CVA{all} revealed clear separation between all 12 fungal groups (fig. 5). The first two CVs in the other five CVAs did not fully separate the groups. However, they each explained more than 65% of the variance in their respective analyses. In addition, while each CV contains all amino acid sites, only a few sites (2–11) contributed to the CVs’ discerning power (supplementary table S2, Supplementary Material online). Thus, overall, only a small number of amino acids were required to discriminate between fungal groups using CVA.
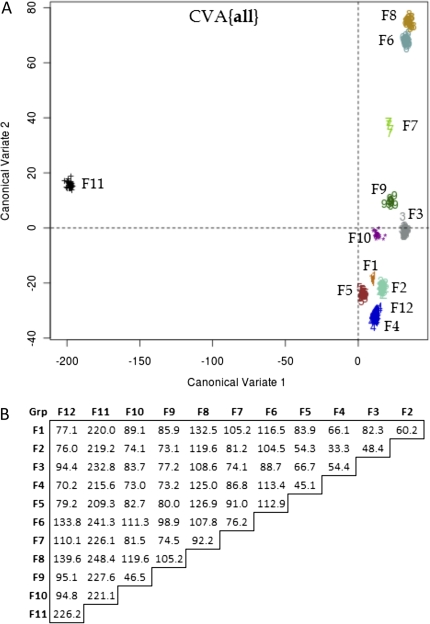
FIG. 5.
CVA of fungal bHLH groups. (A) Projection of 488 fungal bHLH sequences onto eigenvectors (CVs) for the all data set. Plot contains the first and second CV of 11 total. Axes reflect the Mahalanobis distance between fungal groups F1–F12. Group F12 is not discernable from Group F4 with only the first and second CVs. (B) Pairwise Mahalanobis distance between fungal group centroids in the CVA{all} analysis.
Both SWDA and CVA had highly supported (>99% coefficient of agreement) and nearly perfect (>99.9%) accuracies (table 4). CVA and SWDA both determined the sites 6, 8, 10, 11, 12, 15, 50, 51, and 54 (table 2) to be discerning. Site 12 appeared most often in these analyses and was a discerning site for each of the five-factor score data sets in both SWDA and CVA. The other eight sites were found across the six different CVA and SWDA. In summary, using two independent statistical methods, we found nine sites common to both sets of analyses that were central in distinguishing between fungal groups F1–F12.
Simplified Model for F1–F12
To identify the inherent characteristics that effectively separated F1–F12, we utilized the consensus sequence, the decision tree, and the discriminant analyses to manually build a simplified model that characterizes each set of fungal group sequences. As shown in table 5, each fungal group was characterized by a model that used four amino acid sites or less; where groups F4 and F11 were discerned by a single amino acid site (12 and 50, respectively). Site 8 was the most frequently used discerning amino acid site in the model, where amino acids S or A were characteristic of groups F6–F10 and I or V of groups F1 and F3.
Table 5.
Simplified Model for the 12 Fungal Groups.
| Grp | 1 | 4 | 6 | 7 | 8 | 12 | 15 | 16 | 19 | 20 | 50 | 53 |
| F1 | QTM | IV | Y | |||||||||
| F2 | Y | KR | I | |||||||||
| F3 | E | V | ||||||||||
| F4 | Y | |||||||||||
| F5 | S | K | ||||||||||
| F6 | A | R | ||||||||||
| F7 | A | S | ||||||||||
| F8 | L | A | ||||||||||
| F9 | QAL | SA | ||||||||||
| F10 | N | S | I | E | ||||||||
| F11 | E | |||||||||||
| F12 | K | L |
Note.—The bHLH positions and their states (i.e., amino acids) that best distinguish groups F1–F12 are given. Those amino acids in underlined italics are uncharacteristic of a given fungal group (e.g., F2 sequences do not contain Y at site 12).
To assess the effectiveness of the simplified model, we tested it against the sequences from completed fungal genomes that were a priori assigned to a fungal group by our phylogenetic analyses (488 sequences). The simplified model was extremely accurate with a score of 99% and a coefficient of agreement of 98.8% (table 4). Only 8 of the 488 sequences were left unclassified. Thus, the performance of the simplified model to differentiate fungal groups was very similar to the fungal CVA, SWDA, and decision tree analyses.
Classification Model Testing
To test the effectiveness of the different classification models in discerning fungal groups, we determined the sensitivity, specificity, and accuracy for a set of 198 bHLH domains from fungal sequences not used to build the classification models (F.198) (table 4). As shown, all the classification methods had accuracies >92.9% with high coefficients of agreement (>94.7%). Although CVA{all} and SWDA{all} had nearly identical accuracies, SWDA{all} had better performance as it was able to classify 98.4% of the sequences. CVA{all} was only able to classify 90.4% from the F.198 sequence set. The simplified model performed well in both accuracy (96.4%) and sequences classified (195 of 198). However, while the simplified model had great performance, each model tested was extremely accurate for fungal bHLH sequence classification.
Comparison of Positional Amino Acid Conservation
To determine the relationship between fungal and animal sequences, we characterized the conservation of amino acids at specific bHLH positions and compared those patterns with each animal-binding group. Sites 5, 8, and 13 were used by Atchley and Fitch (1997) to classify animal bHLH proteins into either Group A or B. Group A contains an R at site 8, whereas Group B contains amino acids H or K at site 5 and R at site 13. All the fungal sequences fit best into Group B where 94% had an H (0% had a K) at site 5 and 99% had an R at site 13. No fungal bHLH sequences fit the Group A pattern as none had an R at position 8. Animal Group E proteins follow the 5–8–13 Group B rule with the addition of P at site 6. Fungal sequences did not follow this pattern as there were not any sequences with P at site 6. Group C bHLH proteins contain an extra PAS domain, which is not typically found within fungal bHLH proteins. In our data set, the PAS domain was only found in a single protein from S. macrospora. Group F proteins contain the COE domain, not found in fungi. Last, Group D proteins do not bind DNA; however, given the conservation of E at site 9 the vast majority of our sequences are E-box binders. These results support previous studies that fungi bHLH are most closely related to animal Group B.
Phylogenetic Relationship of Fungal bHLH to Animal and Plant bHLH Domains
To further determine the relationship between plant and animal families and our fungal sequences, we built a BA phylogeny based on all sequences from the three Kingdoms. The analysis was based on 916 total sequences, including 147 from Plants, 490 from fungi, and 279 from Animals (six fungal sequences removed). The majority of the previously defined plant, animal, and fungal bHLH groups were identified in corresponding phylogenetic clades with high posterior probabilities (supplementary fig. S1E, Supplementary Material online). Fungal sequences predominantly clustered with animal Group B, however, animal Group B was not conserved as a single clade. This resulted in many previously unidentified evolutionary relationships between Group B and fungal groups F1–F12. For instance, fungal group F2 and four Group B sequences, including TFE3, TFEB, TFEC, and MITF (MiT/TFE family) from H. sapiens, were located in a strongly supported clade. Group F4 was closely related to four animal sequences belonging to the SREB family. These four Group B sequences from Mus musculus, Sus scrofa, and Drosophila melanogaster have biological roles similar to the SRE1 and SRE2 proteins found in fungal group F4. Though interesting, additional comparisons between fungal groups and animal Group B were not supported by high posterior probabilities.
Cross Kingdom Classification
To gain deeper insight into the evolutionary relationship between fungal and animal sequences, we classified 707 fungal bHLH sequences available from Interpro using the animal group classification models described in Atchley and Zhao (2007) (table 6). Every animal model, except the classical animal binding–group model, classified >72% of fungal sequences as animal Group B, with CVA{all} classifying over 88% of fungal sequences as Group B. In most instances, the remaining fungal sequences matched animal Group E. Thus, we found that fungal bHLH sequences were predominately classified as members of animal Group B.
Table 6.
Cross Kingdom Classification of bHLH Domains.
| Group | Decision Tree | CVA {pah} | CVA {pss} | CVA {ms} | CVA {cc} | CVA {ec} | CVA {all} | SWDA {all} | Classic Model |
| A | 0.1 | 0.4 | 18.4 | 8.1 | 3.6 | 0.0 | 4.4 | 0.1 | 0.1 |
| B | 97.4 | 81.2 | 72.0 | 72.7 | 84.4 | 87.8 | 87.8 | 82.4 | 1.4 |
| C | 0.1 | 1.0 | 5.7 | 1.8 | 2.3 | 0.7 | 0.3 | 0.9 | 2.1 |
| D | 2.3 | 0.0 | 0.3 | 0.0 | 0.0 | 0.3 | 0.4 | 0.1 | 0.0 |
| E | 0.0 | 14.4 | 0.7 | 14.4 | 6.8 | 8.3 | 4.1 | 15.9 | 0.0 |
| Unclassified | 0.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 0.6 | 96.4 |
Note.—The percentage of 707 fungal sequences classified into animal Groups (A–E) are reported for animal classification models Decision Tree, SWDA{all}, each CVA, and the Classic model. The percentage unclassified for each model are also reported.
Finally, to determine which groups were most closely related, we calculated the pairwise distances between all fungal and animal groups by building a CVA{all} classification model on the combined F1-F12 and animal Group A–E data sets. Of the 16 CVs (not shown), the first seven explained 94% of the among-groups variation. Animal Groups A, C, D, and E could be separated from each other and all fungal groups within the first four CVs. Additionally, fungal groups F3, F4, F6, and F8–F12 were all distinguishable within the first seven CVs. Group B could not be discerned from the remaining fungal groups until after the seventh CV. The Mahalanobis distance between animal and fungal groups (table 7) supported the close relationship of Group B to fungal groups as Group B had the lowest relative distance from each fungal group averaging 37.6, compared with 121.9 for animal Group D. The average relative distance of fungal to animal groups were much more consistent, with values between 61.5 and 74.1; except F11 with a distance of 129.1. Within this analysis, we also observed that animal Groups B and E were more closely related to each other with a Mahalanobis distance of 29.1, with the other animal pairwise distances ranging from 56.7 to 120.4. Thus, we determined that animal Group B was more similar to fungal groups than any other animal group and there was not a particular fungal group to which animal groups were more closely associated.
Table 7.
Mahalanobis Distance between Animal and Fungal bHLH Groups.
| Grp | A | B | C | D | E | Average |
| F1 | 89.4 | 32.5 | 63.4 | 120.5 | 45.3 | 70.2 |
| F2 | 82.1 | 17.5 | 58.3 | 117.8 | 31.7 | 61.5 |
| F3 | 86.8 | 28.0 | 57.2 | 116.4 | 33.6 | 64.4 |
| F4 | 86.1 | 26.1 | 65.7 | 117.1 | 40.3 | 67.1 |
| F5 | 81.0 | 23.1 | 61.0 | 118.6 | 27.8 | 62.3 |
| F6 | 89.1 | 40.4 | 62.6 | 118.1 | 43.4 | 70.7 |
| F7 | 89.0 | 34.2 | 62.4 | 120.7 | 43.0 | 69.9 |
| F8 | 91.0 | 43.4 | 72.0 | 118.1 | 46.2 | 74.1 |
| F9 | 83.9 | 26.5 | 62.3 | 117.0 | 36.6 | 65.3 |
| F10 | 83.1 | 30.3 | 66.0 | 118.9 | 43.3 | 68.3 |
| F11 | 132.9 | 109.7 | 128.4 | 156.8 | 117.7 | 129.1 |
| F12 | 92.0 | 39.6 | 67.2 | 122.2 | 48.5 | 73.9 |
| Average | 90.5 | 37.6 | 68.9 | 121.9 | 46.4 |
Note.—A CVA{all} was constructed on the entire set of grouped animal and fungal proteins. The relative distance (Mahalanobis distance between group centroids) of F1–F12 and animal Groups A–E are reported. The average of these distances for each fungal and animal group is also shown.
Discussion
Based on the analysis of whole genome projects of fungi, we identified between 4 and 16 bHLH sequences per genome. Overall, the copy count of bHLH proteins is fairly invariant in the Fungal kingdom, with the majority of fungi containing nine bHLH proteins. The bHLH copy count was more consistent within taxonomic groups such as the Onygenales and Eurotiales orders and the Saccharomycetes and Sordariomycetes classes. Thus, the number of bHLH proteins within specific fungal lineages is, in general, strictly conserved. The occurrence of bHLH proteins in Plants and Animals differ dramatically from fungi where Plants contain copy counts of >160 and Animals which have a wider range (50–200) proteins per organism. The lower bHLH copy count, as compared with Animals and Plants, is consistent with but not proportionate to lower gene counts in fungi.
In Ascomycota and Basidiomycota fungi, we identified 12 distinct phylogenetic groups of bHLH domains. Fungal groups F2–F5 and F11 contained representatives with ties to essential biological functions, such as chromosome segregation, interorganelle communication, sexual development, and phospholipid synthesis (table 3). These five groups are found in all fungi examined and were likely present in the MRCA of Ascomycotas and Basidiomycotas.
It is unclear whether 12 fungal groups are linked to specific binding motifs as observed in Animals. Within Animals, the six bHLH groups are linked to specific binding motifs, with the exception of animal Group D, which does not bind DNA. On the other hand, plant bHLH groups are not currently tied to specific binding motifs. Although many of the discerning sites between fungal groups are located in the basic region, this is not always the case. Determination of binding properties of the fungal groups will require additional experimentation.
We observed expansions and losses in all fungal groups, except F3 that had a single representative in every fungal organism. F4 had the largest number of expansions, with at least one set of expansions (subclade A) linked to SREB proteins (table 1, fig. 3). Most fungal organisms were represented at least once in this SREB subclade. This was exemplified by members of the Onygenales, which only had a single F4 sequence. Each was a member of subclade A (data not shown). When evaluating expansions within F4 of Sodariomycete organisms the results favored ancient divergence rather than recent duplication events. Neurospora crassa and other Sodariomycetes exhibit repeat-induced point (RIP) mutation which inactivates duplicated genes (Cambareri et al. 1991; Graïa et al. 2001; Ikeda et al. 2002; Osborne and Espenshade 2009). The presence of these expansions in F4 suggest that these duplications either predate or were protected from RIP. Additionally, F4 was the only group to have the Leucine Zipper domain found across both Basidiomycota and Ascomycota members (fig. 2).
Saccharomycotina organisms appear to have lost the F8 bHLH domain, which has been tied to sexual differentiation and conjugation in Taphrinomycotina organisms (Benton et al. 1993). Additionally, most Saccharomycotina organisms lack the F10 domain, known to be associated with alkane response (Endoh-Yamagami et al. 2007). Likewise, Basidiomycota fungi either lost or never gained the F6 group, which has been linked to phosphate starvation response and chromatin remodeling in Saccharomyces cerevisiae (O’Neill et al. 1996; Then Bergh et al. 2000) as well as copper ion response and sexual/asexual development in N. crassa (Park et al. 2011). Basidiomycota organisms, however, do not lack these biological functions (Morrow and Fraser 2009; Tatry et al. 2009; Mendonça Maciel et al. 2010), possibly utilizing transcription factors with degenerate or missing bHLH domains. As shown in table 3, to date very little is known of the function of bHLH proteins in Fungi. However, we were able to identify that group F3 is associated with chromosomal segregation and several essential biological process within several Pezizomycotina, Saccharyomycotina, and Basidiomycota organisms. Also, we found in Aspergillus fumigatus that the group F5 protein Q6MYV5 is essential in nitrate assimilation and quinate utilization. Thus, bHLH proteins belonging to specific Phylums, Subphylums, and Orders were associated with particular biological functions and conserved motifs. Additionally, many of these associations correlated with bHLH gain and losses within fungal groups.
Saccharomyces cerevisiae bHLH heterodimers YAS1/YAS2 and INO2/INO4 are found in groups F10–F12 with both INO4 and YAS1 in F11. Interestingly, group F10 contains only two Saccharoymycotina sequences, whereas group F12 contains them exclusively. From the phylogenetic analysis, we know that these groups are more closely related to each other than to the other groups (fig. 2). We also know that the relative distance between F10 and F12 is much smaller than either one is to F11 (fig. 5). Given these lines of evidence, it is reasonable to view F10 and F12 as a larger group that is closely related to F11. Thus, the F10/F12 and F11 clades portray the relationship of heterodimers as two distinct yet functionally tied groups. This relationship provides additional insight into potential heterodimers in other Fungi with F10/F12 and F11 bHLH domains.
We built several different models to classify bHLH domains into different groups and determined that fungal group origin could be deduced using only a handful of amino acids. This is very similar to the classical animal binding group model, in that only a few amino acid sites are needed to discern between groups (Atchley and Zhao 2007). Our fungal-simplified model only required 12 amino acid positions to accurately distinguish F1–F12 sequences. In the model, groups F4 and F11 were so distinct that they were identified by a single site. The simple model (table 5) was nearly as accurate as the discriminant analyses (table 4) in testing and was very useful for rapid assessment of fungal bHLH proteins. For example, if a bHLH-containing protein of interest contained a Y at site 12, the simplified model identified it as an F4 sequence. Thus, in many instances, the sequence would be similar to SRE1 and 2 and likely contain an SREB domain.
Many of the most discriminating sites between fungal groups are tied to the fundamental molecular architecture of the bHLH domain, as described primarily with crystal structure studies on animal proteins Max (Ferré-D’Amaré et al. 1993; Brownlie et al. 1997), E47 (Ellenberger et al. 1994), USF (Ferré-D’Amaré et al. 1994), MyoD (Ma et al. 1994), PHO4 (Shimizu et al. 1997), and SREBP (Párraga et al. 1998). For example, site 12 is a highly discerning site useful in identifying group F4 sequences. Site 12 was identified as highly discerning in the decision tree analysis and each of the SWDA and CVAs. It was also used in the simplified model and found to be moderately conserved during the consensus sequence analysis. This site is conserved in animals and has been found to bind the phosphate backbone and/or the DNA within the E-box (De Masi et al. 2011).
Site 50 is another site that is conserved in both Fungi and Animals. It has been determined to pack against buried site 20 and to contact the DNA and/or phosphate backbone in Max, MyoD, PHO4, and USF. In our analyses, it was determined to be a discerning site within the decision tree analysis, significant in many of the SWDA and CVA, and used in the simplified model. From these analyses, we were able to determine that group F11 sequences were uniquely identified by having an E at position 50.
Site 8, known to contact the phosphate backbone and/or DNA of the E-box in MyoD and E47, was a moderately conserved site in fungal sequences. It was the first discerning site in the decision tree analysis and found to be a highly discerning site by both the SWDA{all} and CVA{all}. Site 8 was also utilized in the conventional classification of animal binding groups (Atchley and Fitch 1997) in which amino acids RK were characteristic of animal Group A. However, the fact that it was a discerning site in both models is where the similarity to the animal model ends as RK was not found at site 8 for any fungal sequence.
Use of classification models can find weak linkages, not found using conventional approaches. For example, all members of the Pezizomycotina contained a single copy in F10, except Magnaporthe oryzae. Absence of the F10 bHLH domain was assessed using two methods. Performing a BLAST (Altschul et al. 1990) with several F10 representative domains and a large e-value (10.0) returned only sequences assigned to other fungal groups. A Hidden Markov Model (Bateman et al. 1999) was also constructed from F10 sequences and used to scan the entire M. oryzae genome, with similar results to the BLAST analysis (data not shown).
As shown in table 1, M. oryzae protein MGG_01090 contained an unclassified bHLH domain. However, when we applied the classification models discussed here, we found that four of the nine models classified MGG_01090 as F10 (CVA{pss}, CVA{ec}, SWDA{all}, and the simplified model). The results were not unanimous as the models CVA{ms}, CVA{all} both classified the protein to the closely related F9 group. The decision tree classified MGG_01090 as an F2, which deviated from any of the statistical models. Thus, the developed classification models may be of considerable utility to identify potential group origins of phylogenetic outliers.
Previous work has hinted at a link between the fungal bHLH domain and animal Group B (Atchley and Fernandes 2005; Osborne and Espenshade 2009; Skinner et al. 2010). In our analyses, we provided multiple lines of evidence that Fungi are closely related to Group B. First, in the consensus sequence, it was shown that fungal sequences follow the BxR rule for bHLH positions 5–8–13, characteristic of Group B (Atchley and Fitch 1997). Second, the highest supported clades between Fungi and Animals were only to Group B sequences, specifically linking F2 and F4 to Group B proteins. Third, in our cross kingdom classification analysis, we determined that fungal sequences were predominantly classified into animal Group B. Last, the Mahalanobis distance between Group B and groups F1–F12 was much shorter than any other animal group. Thus, in a comprehensive analysis of fungal bHLH domains, there is clear evidence that fungal sequences are directly related to animal Group B sequences.
We did note that some fungal sequences were classified as animal Group E in the cross Kingdom analysis. The binding domains for Group B and E are very similar. Furthermore, we show that Groups B and E are closely related to each other as evidenced by the Mahalanobis distance between these two groups. However, no fungal sequences contained a P at position 6, required by the classical animal binding group model for animal Group E (Ledent and Vervoort 2001; Atchley and Zhao 2007). Recent studies of Class VI proteins in C. elegans suggest that P is not absolutely required at site 6 to be a member of Group E (Sablitzky 2005; Guimera et al. 2006; Grove et al. 2009). If P is not required, our findings would support that metazoan bHLH sequences may not be uniquely derived from Group B.
In summary, we have determined the conserved sites for the fungal bHLH domain using entropy and consensus sequences. We have also identified 12 major fungal bHLH groups through phylogenetic analysis and tied these groups to conserved domains and biological functions. Using statistical classification models, we have shown that fungal group origin (F1–F12) can be determined with a high degree of accuracy, utilizing only a handful of highly conserved sites that are directly correlated to molecular functions. We have demonstrated the utility of these classification models by identifying group origin with degenerate sequences. Finally, we have made publically available these models, source code, and experimental data at www.fungalgenomics.ncsu.edu.
Supplementary Material
Supplementary data 1, figures S1 and S2, and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We would like to thank Lisa McFerrin and members of the Center for Integrated Fungal Research for their critical comments and discussion. This work was supported by a grant to the Bioinformatics Research Center of North Carolina State University from the National Institute of Health and a grant to R.A.D. from the National Science Foundation (MCB-0731808).
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Fernandes AD. Sequence signatures and the probabilistic identification of proteins in the Myc-Max-Mad network. Proc Natl Acad Sci U S A. 2005;102:6401. doi: 10.1073/pnas.0408964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci U S A. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 1999;48:501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Wollenberg KR, Fitch WM, Terhalle W, Dress AW. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol Biol Evol. 2000;17:164–178. doi: 10.1093/oxfordjournals.molbev.a026229. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Zhao J. Molecular architecture of the DNA-binding region and its relationship to classification of basic helix–loop–helix proteins. Mol Biol Evol. 2007;24:192. doi: 10.1093/molbev/msl143. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton BK, Reid MS, Okayama H. A Schizosaccharomyces pombe gene that promotes sexual differentiation encodes a helix-loop-helix protein with homology to MyoD. EMBO J. 1993;12:135–143. doi: 10.1002/j.1460-2075.1993.tb05639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L, Friedman J, Stone CJ, Olshen RA. Classification and regression trees. 1st ed. Boca Raton (FL): Chapman and Hall/CRC; 1984. [Google Scholar]
- Brownlie P, Ceska T, Lamers M, Romier C, Stier G, Teo H, Suck D. The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control. Structure. 1997;5:509–520. doi: 10.1016/s0969-2126(97)00207-4. [DOI] [PubMed] [Google Scholar]
- Buck MJ, Atchley WR. Phylogenetic analysis of plant basic helix-loop-helix proteins. J Mol Evol. 2003;56:742–750. doi: 10.1007/s00239-002-2449-3. [DOI] [PubMed] [Google Scholar]
- Cambareri EB, Singer MJ, Selker EU. Recurrence of repeat-induced point mutation (Rip) in Neurospora Crassa. Genetics. 1991;127:699–710. doi: 10.1093/genetics/127.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- Endoh-Yamagami S, Hirakawa K, Morioka D, Fukuda R, Ohta A. Basic helix-loop-helix transcription factor heterocomplex of Yas1p and Yas2p regulates cytochrome P450 expression in response to alkanes in the yeast Yarrowia lipolytica. Eukaryot Cell. 2007;6:734–743. doi: 10.1128/EC.00412-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman R, Beran-Steed RK, Anthony-Cahill SJ, Lear JD, Stafford WF, 3rd, DeGrado WF, Benfield PA, Brenner SL. Multiple oligomeric states regulate the DNA binding of helix-loop-helix peptides. Proc Natl Acad Sci U S A. 1993;90:10429–10433. doi: 10.1073/pnas.90.22.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D’Amaré AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D’Amaré AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graïa F, Lespinet O, Rimbault B, Dequard-Chablat M, Coppin E, Picard M. Genome quality control: RIP (repeat-induced point mutation) comes to Podospora. Mol Microbiol. 2001;40:586–595. doi: 10.1046/j.1365-2958.2001.02367.x. [DOI] [PubMed] [Google Scholar]
- Gross ST. The kappa coefficient of agreement for multiple observers when the number of subjects is small. Biometrics. 1986;42:883–893. [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJM. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera J, Vogt Weisenhorn D, Echevarría D, Martínez S, Wurst W. Molecular characterization, structure and developmental expression of Megane bHLH factor. Gene. 2006;377:65–76. doi: 10.1016/j.gene.2006.02.026. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, et al. (37 co-authors) InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Nakayashiki H, Kataoka T, Tamba H, Hashimoto Y, Tosa Y, Mayama S. Repeat-induced point mutation (RIP) in Magnaporthe grisea: implications for its sexual cycle in the natural field context. Mol Microbiol. 2002;45:1355–1364. doi: 10.1046/j.1365-2958.2002.03101.x. [DOI] [PubMed] [Google Scholar]
- Johnson RA, Wichern DW. Applied multivariate statistical analysis. 5th ed. Upper Saddle River (NJ): Prentice Hall; 2001. [Google Scholar]
- Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Ledent V, Paquet O, Vervoort M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002;3:1–18. doi: 10.1186/gb-2002-3-6-research0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, et al. (12 co-authors) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AHK, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;116:843–853. doi: 10.1016/s0092-8674(04)00217-x. [DOI] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, et al. (27 co-authors) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Masi F, Grove CA, Vedenko A, Alibés A, Gisselbrecht SS, Serrano L, Bulyk ML, Walhout AJM. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res. 2011;39:4553–4563. doi: 10.1093/nar/gkr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFerrin L. HDMD: statistical analysis tools for high dimension molecular data [Internet] 2010. [cited 2012 Jan 11]. Available from: http://cran.r-project.org/web/packages/HDMD/. [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–1480. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- Mendonça Maciel MJ, Castro e Silva A, Telles Ribeiro HC. Industrial and biotechnological applications of ligninolytic enzymes of the basidiomycota: a review. Electron J Biotechnol. 2010;13:1–6. [Google Scholar]
- Morgenstern B, Atchley WR. Evolution of bHLH transcription factors: modular evolution by domain shuffling? Mol Biol Evol. 1999;16:1654–1663. doi: 10.1093/oxfordjournals.molbev.a026079. [DOI] [PubMed] [Google Scholar]
- Morrow CA, Fraser JA. Sexual reproduction and dimorphism in the pathogenic basidiomycetes. FEMS Yeast Res. 2009;9:161–177. doi: 10.1111/j.1567-1364.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Nowrousian M, Stajich JE, Chu M, et al. (17 co-authors) De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 2010;6:1–22. doi: 10.1371/journal.pgen.1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill EM, Kaffman A, Jolly ER, O’Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G, Colot HV, Collopy PD, et al. (13 co-authors) High-throughput production of gene replacement mutants in Neurospora crassa. Methods Mol Biol. 2011;722:179–189. doi: 10.1007/978-1-61779-040-9_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Párraga A, Bellsolell L, Ferré-D’Amaré AR, Burley SK. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2009;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2009. [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. (16 co-authors) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rifkin R, Klautau A. In defense of one-vs-all classification. J Mach Learn Res. 2004;5:101–141. [Google Scholar]
- Robbertse B, Reeves JB, Schoch CL, Spatafora JW. A phylogenomic analysis of the Ascomycota. Fungal Genet Biol. 2006;43:715–725. doi: 10.1016/j.fgb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Robinson KA, Lopes JM. SURVEY AND SUMMARY: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 2000;28:1499–1505. doi: 10.1093/nar/28.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sablitzky F. Encyclopedia of life sciences. Chichester (UK): John Wiley & Sons, Ltd; 2005. Protein motifs the helix-loop-helix motif [Internet]. In: John Wiley & Sons, Ltd. [cited 2012 Jan 11]. Available from: http://doi.wiley.com/10.1038/npg.els.0002713. [Google Scholar]
- Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Rawls A, Wilson-Rawls J, Roalson EH. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation. 2010;80:1–8. doi: 10.1016/j.diff.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen GA, Pawlowski L, Wilensky SE, Bender J. Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics. 2002;161:1235–1246. doi: 10.1093/genetics/161.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006;25:3912–3920. doi: 10.1038/sj.emboj.7601270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tatry M-V, El Kassis E, Lambilliotte R, Corratgé C, van Aarle I, Amenc LK, Alary R, Zimmermann S, Sentenac H, Plassard C. Two differentially regulated phosphate transporters from the symbiotic fungus Hebeloma cylindrosporum and phosphorus acquisition by ectomycorrhizal Pinus pinaster. Plant J. 2009;57:1092–1102. doi: 10.1111/j.1365-313X.2008.03749.x. [DOI] [PubMed] [Google Scholar]
- Then Bergh F, Flinn EM, Svaren J, Wright AP, Hörz W. Comparison of nucleosome remodeling by the yeast transcription factor Pho4 and the glucocorticoid receptor. J Biol Chem. 2000;275:9035–9042. doi: 10.1074/jbc.275.12.9035. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoumakas G, Katakis I. Multi-label classification: an overview. International Journal of Data Warehousing & Mining. 2007;3:1–13. [Google Scholar]
- Wang Z, Atchley WR. Spectral analysis of sequence variability in basic-helix-loop-helix (bHLH) protein domains. Evol Bioinform Online. 2006;2:187–196. [PMC free article] [PubMed] [Google Scholar]
- Wollenberg KR, Atchley WR. Separation of phylogenetic and functional associations in biological sequences by using the parametric bootstrap. Proc Natl Acad Sci U S A. 2000;97:3288–3291. doi: 10.1073/pnas.070154797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Castlebury LA, Miller AN, et al. (10 co-authors) An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia. 2006;98:1076–1087. doi: 10.3852/mycologia.98.6.1076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.