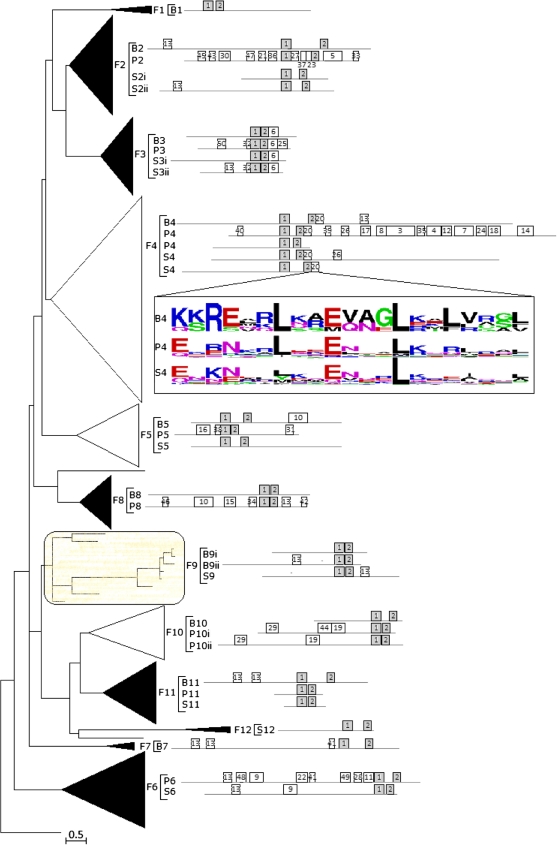
FIG. 2.
Phylogenetic analysis of fungal bHLH. Phylogenetic relationships, taxonomic representation, bHLH motif statistics, and architecture of conserved protein motifs for 12 fungal bHLH groups. ML tree of 490 fungal bHLH proteins (full representation of Basidiomycota, Pezizomycotina, Saccharyomycotina, and Fungal trees available in supplementary fig. S1A–D, Supplementary Material online). The tree is drawn to scale (branch lengths proportional to evolutionary distances) and has been rooted with a single representative from Chlamydomonas reinhardtii. Groups, determined by clades with strong support, are collapsed as triangles with width and depth proportional to the size and sequence divergence of each group, respectively. Groups supported by bootstrap values >30 in NJ or maximum parsimony (MP) analyses are colored black. The shaded group F9 was ambiguously retrieved in NJ, MP, or BA trees. Ungrouped genes are indicated as single lines, and the scale bar represents the estimated number of amino acid replacements per site. Basidiomycota (B), Pezizomycotina (P), and Saccharyomycotina (S) clades associated with each fungal group are noted in brackets. The architecture of conserved motifs, as determined through MEME, is graphically represented as boxes drawn to scale. Box enumeration corresponds to specific motifs found by MEME (supplementary fig. 2C, Supplementary Material online). Grey boxes represent motifs that match the bHLH domain. Last of all, the sequence logo for B4, P4, and S4 for 21 amino acids downstream of the bHLH domain are shown (motif 20).