Figure 4.
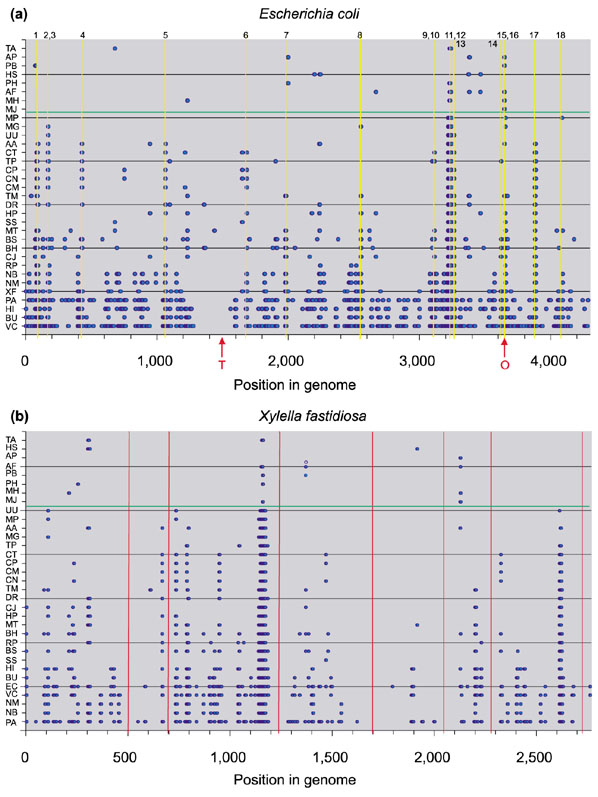
Gene order conservation in the species studied, using (a) Escherichia coli and (b) Xylella fastidiosa as a reference. Position in the reference genome means number of genes from minute zero. Individual species are plotted in the y axis and are ordered according to their phylogenetic distance (estimated by SSU rRNA substitutions) to the reference species. The more closely related species are shown lower down and more distantly related species higher up the axis. Species names are listed in Table 2. Blue dots indicate genes belonging to conserved runs for each species. A horizontal green line separates Bacteria from Archaea. (a) For E. coli, yellow lines show the regions with especially high conservation of gene order. A detailed study of these regions can be found in Table 1. The origin and terminus of replication are marked O and T, respectively, at the bottom of the graph. (b) For X. fastidiosa, red lines indicate regions of high frequency of unique genes [25]. A low degree of gene order conservation was found in these regions.
