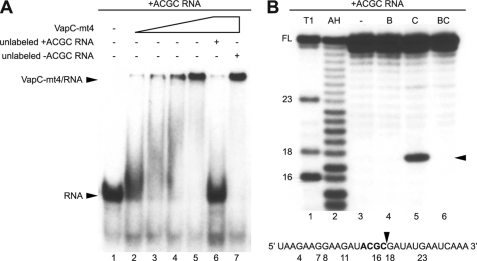
FIGURE 7.
VapC-mt4 selectively binds and cleaves ssRNA containing an ACGC sequence. A, EMSAs were performed in the presence of EDTA, a chelating agent that sequesters Mg2+, to avoid RNA cleavage. His6-VapC-mt4 was able to bind and retard migration of a radioactively labeled synthetic 30-nucleotide RNA containing the ACGC consensus site (lanes 2–5). Sequence specificity was assessed by competition with a 50-fold excess of unlabeled RNA containing (lane 6) or lacking (lane 7) the ACGC consensus site. Data shown are representative of three independent experiments. B, RNA cleavage of the same radioactively labeled synthetic 30-nucleotide RNA containing the ACGC consensus site was performed in the presence of Mg2+, a cation required for VapC-mt4-mediated RNA cleavage. The lane labeled “−” represents a control reaction to which no proteins were added (lane 3). In the lane labeled “B,” purified His6-VapB-mt4 was incubated with the RNA (lane 4). In the lane labeled “C,” the addition of His6-VapC-mt4 resulted in the cleavage of ∼2% of the RNA (lane 5). In the lane labeled “BC,” preincubation of His6-VapC-mt4 with His6-VapB-mt4 prior to the addition of RNA prevented RNA cleavage (lane 6). The cleavage site/product is indicated by a black arrowhead on the right side of the gel and in the relevant RNA sequence below the gel. T1 and AH denote RNA digested with RNase T1 and partially hydrolyzed with alkali, respectively. Numbers indicate the position of G nucleotides. FL indicates the position of full-length RNA. Data shown are representative of two independent experiments.