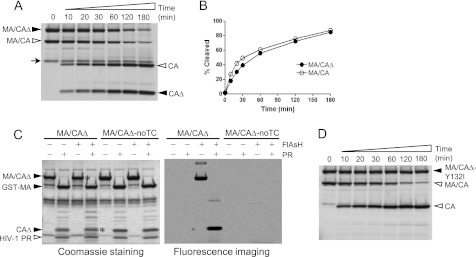
FIGURE 2.
Efficient processing of the MA/CAΔ by HIV-1 PR and specific binding of the FlAsH reagent to the tetracysteine motif. A, both substrates MA/CAΔ and MA/CA were labeled with the FlAsH reagent in the same reaction, and proteolysis of the mixed substrates was performed at 30 °C by adding 0.1 μm HIV-1 PR. Equal molar amounts of the two substrates were used, and the same molar ratio between the FlAsH reagent and total amounts of the protein were used. Aliquots were taken at the various time points shown above each lane and resolved by SDS-PAGE. The aliquot for the 0-min time point was taken before the enzyme was added. Fluorescently labeled proteins were visualized by fluorescence imaging. Filled arrowheads indicate MA/CAΔ substrate and its cleavage product (CAΔ), and open arrowheads indicate MA/CA substrate and its cleavage product (CA). B, the band intensity of the cleavage products, CAΔ and CA, in A was quantified and plotted as a function of the time. C, the substrate MA/CAΔ carrying a tetracysteine motif and the MA/CAΔ-noTC lacking a tetracysteine motif were subjected to labeling reactions in the absence or presence of FlAsH reagent. After the labeling reaction, the proteolysis reaction was performed in the absence or presence of 2 μm HIV-1 PR. The cleavage pattern of the substrates is visualized by Coomassie Blue staining (left) and fluorescence imaging (right). The bands indicated with an asterisk are visualized by fluorescence imaging. D, cleavage of the MA/CAΔ-Y132I is displayed over time to show specific processing of the MA/CA site by the HIV-1 PR.