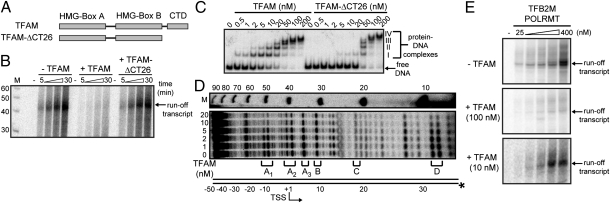
Fig. 3.
Repression of HSP2 transcription by TFAM in vitro. (A) TFAM uses two HMG boxes (Boxes A and B) for DNA binding and CTD for transcriptional activation. The activation portion of the CTD is deleted in TFAM-ΔCT26. (B) TFAM-ΔCT26 does not inhibit runoff transcription from the HSP2-1 template by POLRMT-TFB2M. (C) EMSA of HSP2-1 template over a range of TFAM concentrations shows formation of four complexes (I–IV), with complex I present from 0.5 nM. Formation of complex I is impaired for TFAM-ΔCT26. (D) DNase I footprinting of TFAM-HSP2-1 complexes reveals protection in the regions designated as A1, A2, A3, and D without protection in regions B and C. A schematic of HSP2-1 is shown on the left, and size markers (M) are on the right. (E) Runoff transcription from the HSP2-1 template (100 nM) in the absence or presence of 100 or 10 nM TFAM, which are values that are 10-fold higher than or equivalent to the equilibrium dissociation constant, respectively. In this experiment, product formed at 30 min over the indicated range of POLRMT-TFB2M concentrations.