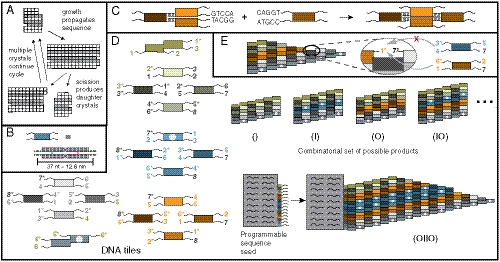
Fig. 1.
Crystal replication. (A) An information sequence (here a column of white or black squares within a 2D crystal) is replicated in two stages. (B–E) DNA tile crystals. (B) DNA tiles are self-assembled, multihelical complexes with 5-nucleotide, single-stranded sticky ends. (C) Tiles attach to crystals via cooperative sticky end hybridization. (D) A tile set that can form ribbon crystals containing any sequence of indigo (I) or orange (O) tile blocks (or the null sequence). Using these tile types, an n-bit wide ribbon could carry any of 2n possible sequences. The two “double” tiles are edge tiles, while the four green tiles below the top edge tile and the four gray tiles above the bottom edge tile, the nucleation barrier tiles, create a thermodynamic barrier to the nucleation of new crystals (25, 24). The four indigo (blue) tiles are the I block tiles and the four orange tiles are the O block tiles. White dots are biotin; colored digits denote sticky end types. Sticky end complements have a *. A crystal seed programmed with sticky ends for a sequence templates growth of that sequence. (E) To be favorable, tiles must attach by at least two sticky ends simultaneously, so growth copies the crystal sequence.