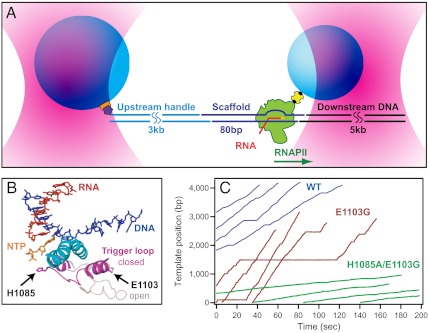
Fig. 1.
Single-molecule assay and transcription records. (A) Experimental geometry for the DNA-pulling optical-trapping assay, based on a “dumbbell” arrangement (not to scale). RNAPII (green) is initiated on a nucleic-acid scaffold (dark blue) that includes template and nontemplate DNA strands plus a short RNA transcript (red). An upstream DNA handle (cyan) and downstream template (black) are ligated to either end of the scaffold. Two polystyrene beads (light blue) are held in separate optical traps (pink). RNAPII is attached to the smaller bead via a biotin:avidin linkage (yellow and black); the DNA handle is attached to the larger bead via a digoxigenin:antibody linkage (purple and orange). Direction of transcription is shown (green arrow); in this orientation, tension assists translocation. (B) Schematic of the RNAPII active site from overlays of two crystal structures, Protein Data Bank (PDB) IDs 2E2H (4) and 1Y1V (41). During transcription, the TL is proposed to fluctuate between a catalytically active “closed” (magenta, 2E2H) and inactive “open” (light pink, 1Y1V) conformation (in this case, restrained by the presence of TFIIS; not displayed). During this transition, the TL comes into proximity with the bridge helix (cyan) and the incoming NTP (orange). The positions of the two TL mutations studied are indicated. (C) Representative records of elongation acquired under 10 pN assisting load for WT RNAPII (blue) and two TL mutant enzymes, E1103G (red) and H1085A/E1103G (green).