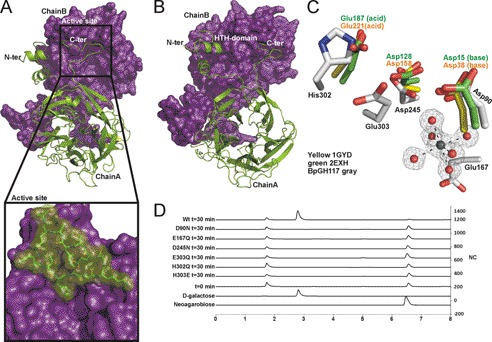
FIGURE 2.
BpGH117 forms dimer in which both monomers contribute to architecture of active site. A and B, the structure of the BpGH117 dimer. Chain A (green) is shown in schematic representation, and Chain B (purple) is shown as a solvent-accessible surface representation. A and the inset show the intimate association of the C-terminal extensions with the neighboring monomer. B shows a slightly rotated view of the dimer to show both the N- and C-terminal interactions as well as the β-propeller fold of the monomer. C, comparison of the active site of BpGH117 (gray) with the catalytic residues in a GH43 arabinase (Protein Data Bank code 1gyd; yellow) and xylanase (Protein Data Bank code 2exh; green) and the proximity of the active site to the magnesium/water cluster. D, high performance anion exchange chromatography with pulsed amperometric detection analysis of neoagarobiose hydrolysis by BpGH117 and various active site mutants. The bottom two traces represent relevant standards, whereas the third trace from the bottom shows a representative time 0 sample before substrate is hydrolyzed. Additional traces show the chromatograms for samples taken after 30 min of enzyme treatment. HTH, helix-turn-helix. NC, nanocoulombs.