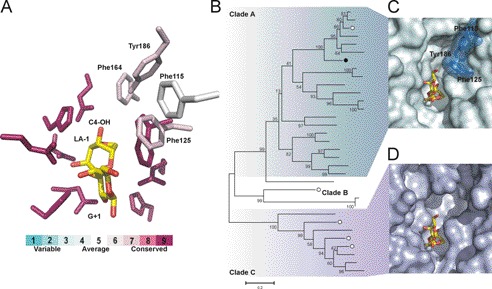
FIGURE 4.
Residues for agarose recognition are highly conserved throughout sequences belonging to glycoside hydrolase family 117. A, the conservation of active site residues (shown as sticks) in GH117 as calculated with ConSurf (see “Experimental Procedures”). The residues that interact with neoagarobiose in BpGH117 are largely conserved in GH117. A cluster of bulky hydrophobic residues that is located at the non-reducing end of the LA residue is less conserved in GH117; these are labeled. B, the phylogenetic tree of GH117 as proposed by Rebuffet et al. (14) and updated here with additional sequences. BpGH117 is indicated by a black circle, and GH117 from Z. galactanivorans is indicated by white circles. The clustering into clades is indicated. (A more detailed phylogenetic tree showing the identity of GH117 members is given in supplemental Fig. 2.) C, the BpGH117 active site is shown with the neoagarobiose complex and the aromatic residues that close the minus (−) subsite at the non-reducing end of the LA residue. BpGH117 belongs to Clade A of the phylogenetic tree, the sequences of which all share the cluster of bulky aromatic side chains at the non-reducing end of the LA residue and thus contain one minus (−) subsite. D, surface representation of a homology model of Zg3615 with neoagarobiose modeled into the active site.