Introduction
I will describe here the way in which I was able to discover that yeast has prions (infectious proteins), proteins that are the carriers of genetic information, and thus are acting as genes (1). This startling finding involved inventing new genetic approaches, but was possible because of my particular background and interests. Our work has broadened the “prion” concept (originally thought to be restricted to a special mammalian disease), established that there are such things as genes made of protein, and led to an understanding of how proteins can encode and transmit heritable information.
My undergraduate degree was from Cornell University in mathematics. This early interest proved important in guiding me into genetics, which I have always viewed as the “logic of life” and, more recently, as I have become involved in solid-state NMR studies. I received an M.D. degree from Georgetown University, and, following a medical internship, I became a postdoctoral fellow with Herb Tabor at the National Institutes of Health (NIH). This was my first real experience doing science, and Herb's critical, careful attitude toward research was my guide in establishing my own approach. I found that adenosylmethionine decarboxylase, the product of which is an intermediate in spermidine biosynthesis, has a pyruvoyl residue as a prosthetic group (2) and that histidine ammonia-lyase has a similar prosthetic group, dehydroalanine (3). As a postdoctoral fellow with Jerry Hurwitz at Albert Einstein College of Medicine in New York, I learned about nucleic acid enzymology from “The Boss” and worked on Escherichia coli DNA polymerase II and in vitro DNA replication (4, 5). I was impressed that the dnats mutants, isolated by Jacob, Bonhoeffer, Carl, Wechsler, and others, were the key to the biochemistry of DNA replication, coupled with in vitro replication systems selected to require the dna gene products (6). In vitro complementation could be used to purify the proteins shown by in vivo studies to be responsible for the biochemical reaction. These experiences having provided me with a firm biochemical background, I then took the 3-week Cold Spring Harbor Laboratory course on yeast genetics, taught by Fred Sherman and Gerry Fink.
I began my independent work in the laboratory of Jerry Hurwitz, who generously allowed me to start working on yeast genetics for a year until my job at the NIH began in 1973. I started out purifying DNA polymerases from yeast, similar to what I had been doing as a postdoctoral fellow with Jerry, but I decided that yeast genetics might be a better route to answering even many biochemical issues. I isolated mutants that could take up dTMP for labeling cellular DNA. These mutants were not widely used for this purpose, but proved quite interesting, particularly tup1 mutants, which had a mating-type α-specific mating defect and a sporulation defect (7) and proved to be a subunit of a transcriptional repressor (8, 9).
The key role that bacteriophage had played in the development of an understanding of bacterial genetics led me to focus on the non-chromosomal genetic element determining the killer trait of yeast (10–12), then newly discovered to involve a double-stranded RNA (dsRNA) in virus particles (13–15). We isolated mutants in host genes necessary for propagation of the killer toxin-encoding M dsRNA (a satellite of the L-A dsRNA virus), mutants that could propagate M dsRNA but could not express the killer phenotype (kex mutants), and superkillers (ski). In an era before yeast cloning, we genetically mapped them as a means of identification.
Major Findings on Yeast Viruses
Our findings on yeast viruses are summarized here. 1) With Steve Sommer, Micheline Wesolowski, and Yutaka Matsumoto, we found there are at least four independent RNA replicons in many strains, namely the L-A virus (4.6 kb, dsRNA), the L-BC virus (also 4.6 kb, dsRNA), and the T and W dsRNAs, forms of the 20 S and 23 S single-stranded RNA replicons (16–19). The latter two replicons have been studied in depth by Rosa Esteban and Tsutomu Fujimura in Salamanca, Spain. 2) Natural variants of the L-A virus interfere with each other and vary in their ability to propagate the M dsRNA satellite RNAs (20, 21). 3) Rosa Esteban and I demonstrated “head-full replication” of the dsRNAs, with intraviral replication until the viral head is full and then extrusion of new (+)-strands for translation and formation of new particles (22). 4) Juan Carlos Tercero and I found a new protein N-acetyltransferase (Mak3p) whose N-terminal acetylation of the major L-A coat protein is necessary for virus assembly (23). 5) Yasuyuki Ohtake and I showed that the level of 60 S ribosomal subunits is critical for L-A virus and particularly M satellite propagation (24), and Herman Edskes showed that this is probably because the viral mRNAs lack poly(A) (25). 6) Akio Toh-e, Steve Sommer, Porter Ridley, and Lionel Benard found seven genes that we named SKI genes (for superkiller) and that Dan Masison and Bill Widner showed limit the expression of virus information by limiting the expression of non-poly(A) mRNAs (such as the viral mRNAs) (26, 27). These genes have homologs in all eukaryotes, and we showed they are critical for preventing viral pathology (27–31). In fact, Anji Searfoss showed that yeast cells deleted for ski2 and its homolog, slh1, translate poly(A)− and poly(A)+ mRNAs with equal efficiency and the same kinetics (32). The translation apparatus does not really need the 3′-poly(A) on the mRNA. 7) Tsutomu Fujimura established in vitro packaging, replication, and transcription systems for L-A, the first for a dsRNA virus, and, with Rosa Esteban and later with Juan Carlos Ribas, we used these systems and genetic experiments to define the RNA sites determining genome packaging and replication and the protein domains required for these processes (33). Much of this work provided a model for later work on mammalian dsRNA viruses (34). 8) Tateo Icho and Tsutomu Fujimura showed that the L-A virus expresses a gag-pol fusion protein (35, 36), and Jon Dinman showed that this occurs by ribosomal frameshifting, much like retroviruses (37). Jon's detailed characterization of this process defined the RNA signals controlling frameshifting and the influence of various genes (38, 39), and he has gone on to extensively explore this area (40, 41). 9) Simple size and density measurements by Rosa Esteban showed that the L-A virus has ∼120 subunits per particle. This was supposed to be a forbidden symmetry, but we suggested that the virus had T = 1 symmetry with an asymmetric dimer as the unit element (22). Cryo-EM studies (42) and the x-ray diffraction structure of the particles obtained by Jack Johnson's group (43) showed that our speculation was correct. It proved to be true of the cores of mammalian dsRNA viruses as well. 10) Our kex1 and kex2 (killer expression) mutants, which were unable to produce active killer toxin or α-pheromone (44), led to the discovery by others of homologous enzymes responsible for processing insulin and other prohormones in mammals (45, 46).
Pathway to Discovery of Yeast Prions
Two aspects of the virus system put us in a position to discover yeast prions. The structure and biochemistry of the L-A virus and the M satellite dsRNA were the same as those of the cores of mammalian dsRNA viruses, and yet the yeast viruses were not able to leave one infected cell and enter another except by artificial means. The yeast viruses spread horizontally when infected cells mate with uninfected cells, and the meiotic progeny all have the viruses. In fact, there are animal and plant viruses that propagate in this way. We thus expected that a yeast infectious protein would have the same property of appearing as a non-chromosomal genetic element. The second point, which was really central, was that our extensive studies of the mak mutants, which were unable to propagate the killer toxin-encoding M dsRNA, prepared us to expect that such mutants would (of course) have a phenotype due to the loss of the M dsRNA (lack of toxin production), a phenotype opposite to that of the presence of the M dsRNA. Thus, when we saw in the literature a non-chromosomal genetic element with a phenotype the same as that of a recessive mutant in a chromosomal gene necessary for the propagation of the non-chromosomal element, we knew that there must be an interesting explanation.
Original Description of [PSI+] and [URE3] by Cox and Lacroute
In 1965, Brian Cox described a non-chromosomal genetic element ([PSI+]) that enhanced a weak nonsense suppressor, SUQ5 (47). He used the ade2-1 mutation, a premature UAA terminator, which resulted in the accumulation of a red pigment derived from the Ade2p substrate, phosphoribosylaminoimidazole. SUQ5 proved to be a serine-inserting tRNA mutant that reads the UAA codon, albeit poorly (48). When the [PSI+] genetic element was also present, the read-through of the premature UAA stop codon in the ade2-1 gene was efficient enough to allow cell growth without adenine.
In 1971, while studying uracil biosynthesis, Francois Lacroute found that cells with a block in the first step, aspartate transcarbamylase (ura2), could not grow on the product of that enzyme, ureidosuccinate. When he isolated cells that could use ureidosuccinate, he found chromosomal mutants, which he named ure1 and ure2, and a non-chromosomal mutant, which he designated [URE3] (49). With Lacroute, Michel Aigle made the key finding that mutants in ure2 could not propagate the [URE3] genetic element (50)! Thus, recessive ure2 mutants with the same mutant phenotype as the presence of the [URE3] element could not propagate [URE3]. I saw that this relationship was different from that of the many mak mutants I had isolated and studied with the killer toxin-encoding M dsRNA. But what did that relationship mean?
Properties of [URE3] Suggested That It Is a Prion of Ure2p
In 1989, when I began these studies, I was aware that the infectious agent of the mammalian transmissible spongiform encephalopathies (TSEs) had been suggested to be an infectious protein (51–53), although the evidence for that conclusion was far from complete, and there was a great deal of disagreement in the field. Indeed, some of the data thought to be strongly supportive of the “protein-only” model proved later to have other explanations. On discovering prion protein (PrP) as the main protein constituent of the purified infectious agent, Prusiner coined the term prion to represent such an entity (53).
It was clear to me that a yeast prion would have properties quite different from those of the mammalian TSEs. In the mammalian disease, accumulation of a toxic form of the PrP protein in the non-growing central nervous system was believed to produce the fatal TSEs. Because yeast is often growing (at least under laboratory conditions), it seemed to me then that accumulation of something toxic would not likely be a problem (but see below). In thinking what properties one would expect of a yeast prion, the prion's being lethal or benign seemed to me to be irrelevant, just as viruses can be benign or lethal. Rather, I sought genetic features that would not depend on the particular phenotype produced by the prion.
The fact that the URE2 gene is necessary for the [URE3] non-chromosomal genetic element and yet ure2 mutants have the same phenotype as the presence of [URE3] (50) struck me as just what one would expect of a prion of Ure2p. Either the ure2 mutation or the conversion to the prion form (whatever that may be) should inactivate Ure2p and produce the same or a similar phenotype in each case. This became the first of my “genetic criteria for a yeast prion” (Fig. 1) (1). Note that this is not a property of the mammalian prion. Deletion of the gene encoding PrP produces no clear phenotype, although the presence of the prion (a TSE) is uniformly lethal. Interestingly, although not mentioned in the published paper that I read (50), Michel Aigle, in his master's thesis, inferred part of the prion explanation of [URE3] to explain his result: “Le facteur [URE3] rendrait donc non fonctionnel le produit du gene (URE 2-+).” (The [URE3] factor thus rendered nonfunctional the product of the URE2 gene.)
FIGURE 1.
Three genetic criteria for a prion of yeast (1). I proposed that these properties are expected of prions but not of nucleic acid replicons (viruses or plasmids) and so should allow diagnosis of yeast prions among non-chromosomal genetic elements.
I tried to think what other genetic properties would distinguish a prion of yeast from a nucleic acid replicon. Bacterial plasmids are often curable by intercalating agents, and yeast mitochondrial DNA is efficiently cured by growth on ethidium bromide. The dsRNA viruses are cured by growth of cells at elevated temperatures. Thus, a given curing agent may or may not cure a nucleic acid replicon, but, for each of the above cases, the cured cells stay cured unless the virus or plasmid is introduced from elsewhere, by mating, transformation, or, in some bacteria, conjugation. In contrast, a prion could arise again spontaneously in the cured strain at some low frequency because the protein capable of conversion to the self-propagating altered form is always present in the cells (1). I called this “reversible curability” (Fig. 1). The [PSI+] prion had been shown to be cured by growth in high osmotic media (54), but cells cured in this way could again become [PSI+] (55). I tested whether [URE3] could be cured by guanidine, as previously reported for [PSI+] (56), and it was indeed cured (1). From the cured strains, I could isolate [URE3] derivatives at frequencies similar to those at which they arose originally (1), so [URE3] satisfied this genetic criteria for a prion.
Because a prion must be based on a self-propagating change, the generation of the prion form in a cell should take over the population of molecules of the prion protein. I surmised that the larger the population of prion proteins, the more frequently the prion change should occur (Fig. 1) (1). The gene for the prion protein should, of course, be a gene needed for the propagation of the prion. Plasmids and viruses of yeast also will depend on chromosomally encoded proteins for their propagation, but, in contrast to the prion case, overproduction of one of those proteins would not induce the de novo generation of the virus or plasmid. This approach provides (a) evidence that a particular non-chromosomal genetic element is a prion, (b) a concrete means for generating the prion formation, and (c) a way to find which protein encodes a given putative prion protein. Indeed, when I tested whether the frequency of [URE3] arising was enhanced by overproduction of Ure2p, I found a robust 100–200-fold effect (1). I can still remember my excitement when this experiment turned out just the way I had expected.
None of my three genetic criteria for a yeast prion (Fig. 1) are known to be true of mammalian prions. There is no way to cure the TSEs, and, even if there were, there would be no way of demonstrating spontaneous generation of mammalian prions in a wild-type mouse because it would be too rare. Overexpression of PrP is known to kill mice (57), but their tissues are not infectious, so prions have not been generated. Deletion of the Prnp gene encoding PrP has no phenotype, unlike the lethal phenotype of a TSE.
Reinterpretation of the Old [PSI+] and [URE3] Literature
The fact that [URE3] satisfied all three genetic criteria for a prion of Ure2p led me to examine whether other orphan non-chromosomal genetic elements could also be prions (1). [PSI+] had been extensively studied, mostly by its discoverer, Brian Cox, and his colleagues, particularly Mick Tuite, and also by Fred Sherman and Sue Liebman. Michael Ter-Avanesyan's group in Moscow and Yury Chernoff, then a student of Sergey Inge-Vechtomov and later a postdoctoral fellow with Bun-Ichiro Ono and Sue Liebman, did key experiments.
As mentioned above, Singh and Sherman had found that high osmotic strength cured [PSI+], but Lund and Cox found that [PSI+] clones could be isolated from a cured strain. On overproduction of Sup35p, the frequency of [PSI+] arising was dramatically increased (58). Finally, [PSI+] propagation depends on the SUP35 gene (59), but the phenotype of [PSI+] and that of sup35 mutants are similar. The original sup35 mutants were recessive “omnipotent suppressors,” meaning that they would suppress UAA, UAG, or UGA nonsense codons (60, 61). We reinterpreted these results as evidence that [PSI+] was a prion of Sup35p (1). Subsequently, our “genetic criteria” have been central to the identification of other yeast and fungal prions (62–66). For example, [Het-s] is a prion of the HET-s protein from Podospora anserina involved in heterokaryon incompatibility (62), and [PIN+] is a prion of yeast Rnq1p (63). The dominance of [URE3] over its absence (denoted [ure-o]) or the dominance of [PSI+] over [psi−] would not mean that the prion form is active in regulating nitrogen catabolism or translation termination, rather that the prion form inactivates the active form by converting it into the prion form.
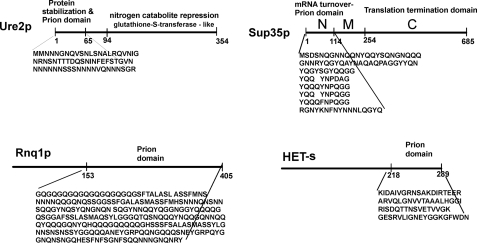
We found that the overproduction of just the first 65 residues of Ure2p was sufficient to induce de novo generation of [URE3] and was actually ∼100 times more efficient than the full-length protein in doing so (Fig. 2) (67). The same fragment was also sufficient to propagate [URE3] in the complete absence of the remainder of the protein (68). We named this the “prion domain.” Earlier elegant studies by Michael Ter-Avanesyan and colleagues showed that the N-terminal 114 amino acid residues of Sup35p are dispensable for growth but necessary for propagation of the [PSI+] prion (69); we reinterpreted this as the Sup35p prion domain (Fig. 2) (67).
FIGURE 2.
Domain structure of yeast and fungal prion proteins. The N-terminal domains of Ure2p and Sup35p have cellular functions, constitute the portion of the molecule necessary and sufficient for prion propagation, and comprise the core of the amyloid.
[PSI+] and [URE3] Prions Are Based on Amyloid Forms of Sup35p and Ure2p
Although the genetic criteria provided strong evidence that [PSI+] and [URE3] are prions of Sup35p and Ure2p, respectively, they did not provide clues to the mechanism of prion action, how a protein could transmit an infection. Dan Masison, in my group, obtained the first biochemical evidence for yeast prions in showing that a fragment of Ure2p is protease-resistant specifically in [URE3] prion-containing cells (67), reminiscent of the remarkable protease resistance of PrP in brains of animals infected with a TSE (53), a reflection of the amyloid form of PrP in the disease. Amyloid is a filamentous polymer of protein monomers composed of β-strands perpendicular to the long axis of the filaments (70). Amyloids of various proteins are also key to the pathogenesis of Alzheimer disease, Parkinson disease, and amyotrophic lateral sclerosis, and amyloid is important in adult-onset diabetes mellitus. Yury Chernoff, in Sue Liebman's laboratory, found that Hsp104p, a disaggregating chaperone, was critical for the propagation of the [PSI+] prion, suggesting that protein refolding might be important in prion formation (71).
Paushkin et al. (72) showed that the self-propagating inactivation of Sup35p in vivo was paralleled by the self-propagating aggregation of Sup35p in vitro and were first to suggest that the role of Hsp104p in prion propagation was the breakup of aggregates to make new seeds. King and Wüthrich (73) were first to show that a yeast prion domain (of Sup35p) could form amyloid in vitro, and Glover et al. (74) showed that extracts of [PSI+] cells could seed amyloid formation by Sup35NM, including the prion domain. Herman Edskes showed that Ure2p is aggregated in [URE3] cells (75). Moreover, Kim Taylor found that Ure2p forms amyloid in vitro, and, just as the Ure2p prion domain is necessary for in vivo prion formation, it is required for in vitro amyloid formation (76). The Ure2p prion domain that can initiate prion formation in vivo can likewise speed amyloid formation by the full-length protein in vitro (76).
The ease of yeast genetics made possible convincing evidence of yeast prions as described above, but, in mammals, some of these approaches were impossible. As a result, attempts to infect animals with amyloid made of recombinant PrP protein has been a critical goal. This was first achieved for the fungal prion [Het-s] (described in more detail below) by Marie-Lise Maddelein in Sven Saupe's group, who showed that amyloid of recombinant HET-s protein efficiently infected fungal cells with the [Het-s] prion, but that the soluble form, or non-amyloid aggregates, did not (77). It was then shown that [PSI+] (78, 79) and, later, [URE3] and [PIN+] (80, 81) could also be infected with amyloids of the prion domains of Sup35p, Ure2p, and Rnq1p.
[β], a Prion Unrelated to Amyloid Formation
The definition of prion is “infectious protein” and does not require a role for amyloid formation (53). We showed that the self-processing vacuolar protease B of yeast (Prb1p) could be the basis for a prion. Prb1p is made as an inactive precursor that is normally processed by protease A (Pep4p) to form the mature active form (82). Zubenko et al. (83) showed that active Prb1p could also activate the Prb1 precursor in the absence of protease A. Tibor Roberts and I showed that this self-activation could continue indefinitely under conditions that derepress Prb1p synthesis and that this phenomenon has all the properties of a prion (64). In this case, the active protease B is the prion form.
Protein Sequence Versus Amino Acid Content in Determination of Prion-forming Ability
A close similarity of prion protein sequence in donor and recipient has long been known as a critical condition for transmission of prion infection; in some cases, even a single amino acid difference can prevent transmission (59, 84). This is the extreme case of the “species barrier,” the delayed or blocked transmission of infection from one species to another (e.g. Ref. 85). In some cases, the altered sequence can form a prion on its own, even though it cannot be infected by the prion from the wild-type sequence (e.g. Ref. 86).
With the intention of proving that the sequence of the Ure2p prion domain was important for prion formation, Eric Ross, in my laboratory, replaced the normal prion domain with each of five randomly shuffled versions of this region in which the amino acid content was preserved but the sequence was randomized (87). Surprisingly, each of these shuffled domains could support de novo prion formation, and he obtained the same result for the prion domain of Sup35p (88). These results showed that, at least for these prion domains, it is the amino acid content, not the sequence, that determines the ability to form a prion. Ross has pursued this area independently and determined what compositional components are critical for prion formation and used this information to create a “designer prion” (89, 90).
Yeast Prions Are In-register Parallel Amyloids: Structure Explaining Biology
From the prion domain shuffling results, we inferred that the amyloid underlying these yeast prions must have an in-register parallel architecture (91). The requirement for sequence near-identity for prion propagation implies a favorable interaction between amino acid side chains. If this interaction were between different, perhaps complementary side chains (like the complementarity of DNA strands), shuffling the prion domain sequence would destroy the complementarity (as it does the hybridization of complementary DNA chains), but if the interaction were between identical side chains (in an in-register parallel structure), shuffling the sequence would leave the side chains still able to have the favorable interactions, just in a different sequence.
To test this reasoning, Frank Shewmaker, Ulrich Baxa, and I collaborated with Rob Tycko, an outstanding solid-state NMR expert who had already shown that amyloid fibers of amyloid β, the central pathogenic element of Alzheimer disease, is an in-register parallel β-sheet structure (92). The high infectivity of amyloid formed in vitro from recombinant yeast and fungal prion proteins, not yet possible with mammalian prion protein, meant that we were studying the structure of the right stuff. A dipolar recoupling experiment, which measures the distance from one 13C-labeled atom to the next nearest labeled atom, can be used to distinguish the in-register parallel architecture from antiparallel, β-helix, or parallel out-of-register structures. Molecules are labeled at one or a few carbonyl carbons, and, in the in-register parallel case, this distance will be ∼4.8 Å, the distance between adjacent β-strands (93). For the other architectures, the distance will generally be twice 4.8 Å or greater. Using highly infectious amyloid of the prion domains of Sup35p, Ure2p, and Rnq1p labeled with the α-carbonyl carbon in any of several different amino acid residues, we consistently found a distance of ∼5 Å in such experiments (94–96). Making amyloid fibers from a mixture of labeled and unlabeled molecules showed that the nearest neighbor was on a different molecule, as expected for the in-register parallel structure. Frank Shewmaker and I showed that filaments of Sup35p conferring different prion variants on infection into yeast each had this architecture (97), and the shuffled prion domains had the same architecture, as predicted (98). This adventure into NMR was especially satisfying to me because it gave me an excuse to pursue my long-standing interest in math and physics on company time. Rob Tycko was particularly generous of his time and instruction, not only doing many of the experiments but also teaching me some of the basics and letting me use his NMR spectrometers.
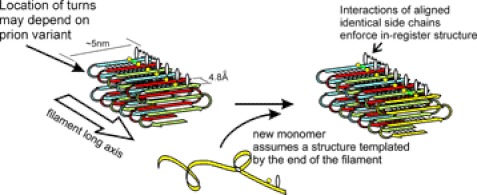
Measurements of mass per unit length of Ure2p and Sup35p amyloid fibers show a single molecule per ∼4.7 Å, consistent with the in-register parallel architecture (Fig. 3) (99–101) but, significantly, ruling out a β-helix (such as is found in the HET-s amyloid (102)) or several other possibilities. The in-register parallel architecture determines much of the structure, but, if the entire prion domain of even the ∼65-residue Ure2p prion domain were in a single flat sheet, filaments would be >20 nm wide. In fact, they are only ∼4–5 nm wide, indicating that the sheet must be folded several times lengthwise, as diagrammed in Fig. 3. We hypothesized that different prion variants have these folds in different locations (103–105).
FIGURE 3.
The in-register parallel β-sheet architecture with longitudinal folds of yeast prion amyloid filaments can explain transmission of conformational information to monomers joining the end of the filament. The same interactions among aligned identical amino acid side chains that hold the structure in-register also drive the monomer joining the end of the filament to have the same conformation as the molecules already in the filament (103, 106).
How Does a Protein Template Its Own Conformation?
The fact that any of a variety of TSE variants (“strains”) can propagate in mice with the same PrP protein sequence made many skeptical of the prion hypothesis because no mechanism for propagation of conformation was even hypothesized. We have proposed that the same positive interactions between identical amino acid side chains that maintain the structure in-register also drive the monomer joining the end of the filament into the same conformation as the monomer already on the end of the filament and, thus, as all the other molecules in the filament. These positive interactions, hydrogen bonds between the side chains of aligned glutamine, asparagine, serine, or threonines residues or hydrophobic interactions among aligned leucine, valine, isoleucine, tyrosine, or phenylalanine residues, are possible if the sequences are aligned, but not if they are off by even a single residue. The turns of each strand will be forced to occur at the same point on the peptide chain in each new molecule joining the end of the filament (Fig. 3). In this way, a protein can template its own conformation, just as a DNA strand can template its own sequence (103–106).
Biology of Yeast Prions: a Help or a Hindrance?
The description by Sven Saupe and coworkers in Bordeaux, France, of a prion, [Het-s], from the filamentous fungus P. anserina involved in a normal fungal function, heterokaryon incompatibility (62), led me to suggest that this was the first case of a potentially beneficial prion (107). However, later work showed that [Het-s] is also involved in a meiotic drive phenomenon, detrimental to the host but promoting the spread of the het-s allele encoding the prion protein (108). Perhaps both are driving the evolution of het-s, but it seems certain that the HET-s protein has evolved to be a specific prion.
The commonly used strains with [PSI+] or [URE3] appear to grow well in the laboratory, but perhaps they are commonly used for this reason, and no consistent reproducible benefit of being [PSI+] or [URE3] has been reported. To address the benefit versus detriment issue, we reflected that even lethal prions are often found in nature (scrapie and chronic wasting disease) because their infectivity outruns the damage they do to their hosts. A beneficial prion, for which infectivity and effect on the host would be working together to promote its spread, instead of in opposition, would quickly become nearly universal in the wild. Thus, a prion that is not found in the wild must be a substantial detriment to its host. Toru Nakayashiki and I surveyed 70 wild-type strains and found each of the known parasitic non-chromosomal nucleic acid replicons (viruses and plasmids) in various proportions of the strains, but none carried the [PSI+] or [URE3] prion (109). This is a sort of sum over all conditions and tells us that the net result of these prions is detrimental.
Dan Masison's group showed that infection of yeast by the [URE3] and [PSI+] prions induces the increased expression of stress proteins Hsp104 and Hsp70s, indicating that the cell views them as detrimental (a stress) (110, 111). Our naming the regions responsible for prion formation “prion domains” may have been an unfortunate choice, as it apparently made others think that this is the function of these domains. In fact, the prion domain of Sup35p has a normal non-prion role in promoting normal turnover of mRNAs by interacting with the poly(A)-binding protein and with the poly(A)-degrading nucleases (112–114). Frank Shewmaker showed that the Ure2p prion domain is required for the normal stability of the protein against degradation, so deletion of this part results in a partially defective phenotype (115). Calling these parts of Ure2p and Sup35p the “prion domains” might be like calling the humerus the “broken arm domain.”
The sequence conservation of the Ure2p and Sup35p prion domains is probably to conserve the non-prion functions of these domains (see above) and is thus not an argument that prions must benefit the host. Indeed, as discussed above, sequence need not be conserved to preserve prion-forming ability in these cases. Moreover, prion-forming ability is not in fact generally conserved. For example, the N-terminal domain of the Candida glabrata Ure2p is similar in sequence to the Saccharomyces cerevisiae Ure2p prion domain, but it cannot form a prion (116, 117). In contrast, the N-terminal domain of the Candida albicans Ure2p is much less similar to that of S. cerevisiae, but readily forms a prion (116). In addition, Ure2p of Kluyveromyces lactis and that of Saccharomyces castellii are each unable to form prions (118, 119).
Scrapie, the prion disease of sheep, has been common in the West for centuries (120) and perhaps for millennia in the East (121). Probably as a result, there have arisen alleles of sheep PrP that are resistant to scrapie infection (122). The incidence of human TSE is limited by a polymorphism at residue 129, which can be Met or Val. Homozygotes of either Met/Met or Val/Val can have Creutzfeldt-Jakob disease, but heterozygotes rarely develop the disease (123). David Bateman found that polymorphisms in the N and M domains of Sup35p likewise limit the spread of [PSI+] in S. cerevisiae (124). We suggest that these polymorphisms became widespread because [PSI+] is not good for yeast.
Although the commonly studied variants of [URE3] and [PSI+] grow well, we suspected that there could easily be [PSI+] variants that soak up all of the Sup35p into amyloid filaments and thus be lethal. Ryan McGlinchey detected such “suicidal” [PSI+] variants, as well as very sick variants constituting over half of all [PSI+] isolates (125). Dmitry Kryndushkin found frequent [URE3] isolates that produce extremely slow cell growth in a background in which deletion of URE2 does not slow growth at all, indicating that this prion as well can be devastating, but by a toxic mechanism, not simply depletion of Ure2p (125). Certain mutants in SIS1, encoding an Hsp40 protein, make even the usual mild [PSI+] variants become lethal to the cell, again emphasizing the danger of being [PSI+] (126). These results show that the impact of acquiring either of these prions is not as benign as previously thought.
Thus, a variety of lines of evidence indicate that the yeast prions [URE3] and [PSI+] are detriment to their hosts. It remains possible that some variant of one of these prions or another prion will be found to be beneficial under some condition. However, it is already clear that these two prions are detrimental on the whole.
Prospects for Future Work on Yeast Prions
It is very satisfying to me that my work on the relatively obscure yeast killer virus led to our discovery of protein-based inheritance, a concept of broad interest. As models for the mammalian TSEs, yeast prions have dramatically advanced our understanding of what a prion can be and how information may be encoded in a protein and even provided the most convincing evidence that there can be such a thing as a prion. Recently, work on other mammalian amyloidoses has advanced the notion that many involve similar mechanisms of spread within the body, that several are transmissible by injection, and that a few may even be naturally infectious (reviewed in Refs. 127 and 128). Unlike the rare TSEs, these amyloid diseases are widespread, and it is clear that the yeast prion systems can provide important information with a wide application in their study. We expect that studies of the mechanisms by which yeast prions are generated, propagate, and produce pathology in their hosts will continue to produce insights important for the wider field of amyloid diseases.
Acknowledgments
I am grateful to all of the bright and dedicated people who have worked with me over the years. I also want to express special thanks to Herb Tabor, who got me started in research, and to my long time collaborator, Herman Edskes.
REFERENCES
- 1. Wickner R. B. (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569 [DOI] [PubMed] [Google Scholar]
- 2. Wickner R. B., Tabor C. W., Tabor H. (1970) Purification of adenosylmethionine decarboxylase from Escherichia coli W: evidence for covalently bound pyruvate. J. Biol. Chem. 245, 2132–2139 [PubMed] [Google Scholar]
- 3. Wickner R. B. (1969) Dehydroalanine in histidine ammonia-lyase. J. Biol. Chem. 244, 6550–6552 [PubMed] [Google Scholar]
- 4. Wickner R. B., Wright M., Wickner S., Hurwitz J. (1972) Conversion of φX174 and fd single-stranded DNA to replicative forms in extracts of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 69, 3233–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wickner R. B., Ginsberg B., Hurwitz J. (1972) Deoxyribonucleic acid polymerase II of Escherichia coli. II. Studies of the requirements and the structure of the deoxyribonucleic acid product. J. Biol. Chem. 247, 498–504 [PubMed] [Google Scholar]
- 6. Schaller H., Otto B., Nüsslein V., Huf J., Herrmann R., Bonhoeffer F. (1972) Deoxyribonucleic acid replication in vitro. J. Mol. Biol. 63, 183–200 [DOI] [PubMed] [Google Scholar]
- 7. Wickner R. B. (1974) Mutants of Saccharomyces cerevisiae that incorporate deoxythymidine 5′-monophosphate into deoxyribonucleic acid in vivo. J. Bacteriol. 117, 252–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams F. E., Varanasi U., Trumbly R. J. (1991) The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol. Cell. Biol. 11, 3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. (1992) Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68, 709–719 [DOI] [PubMed] [Google Scholar]
- 10. Makower M., Bevan E. A. (1963) The inheritance of a killer character in yeast (Saccharomyces cerevisiae). Proc. Int. Congr. Genet. XI 1, 202 [Google Scholar]
- 11. Somers J. M., Bevan E. A. (1969) The inheritance of the killer character in yeast. Genet. Res. 13, 71–83 [DOI] [PubMed] [Google Scholar]
- 12. Fink G. R., Styles C. A. (1972) Curing of a killer factor in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 69, 2846–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bevan E. A., Herring A. J., Mitchell D. J. (1973) Preliminary characterization of two species of dsRNA in yeast and their relationship to the “killer” character. Nature 245, 81–86 [DOI] [PubMed] [Google Scholar]
- 14. Vodkin M., Katterman F., Fink G. R. (1974) Yeast killer mutants with altered double-stranded ribonucleic acid. J. Bacteriol. 117, 681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herring A. J., Bevan E. A. (1974) Virus-like particles associated with the double-stranded RNA species found in killer and sensitive strains of the yeast Saccharomyces cerevisiae. J. Gen. Virol. 22, 387–394 [DOI] [PubMed] [Google Scholar]
- 16. Sommer S. S., Wickner R. B. (1982) Yeast L dsRNA consists of at least three distinct RNAs; evidence that the non-Mendelian genes [HOK], [NEX], and [EXL] are on one of these dsRNAs. Cell 31, 429–441 [DOI] [PubMed] [Google Scholar]
- 17. Wesolowski M., Wickner R. B. (1984) Two new double-stranded RNA molecules showing non-Mendelian inheritance and heat inducibility in Saccharomyces cerevisiae. Mol. Cell. Biol. 4, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsumoto Y., Fishel R., Wickner R. B. (1990) Circular single-stranded RNA replicon in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 87, 7628–7632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rodriguez-Cousiño N., Esteban L. M., Esteban R. (1991) Molecular cloning and characterization of W double-stranded RNA, a linear molecule present in Saccharomyces cerevisiae. Identification of its single-stranded RNA form as 20 S RNA. J. Biol. Chem. 266, 12772–12778 [PubMed] [Google Scholar]
- 20. Wickner R. B. (1980) Plasmids controlled exclusion of the K2 killer double-stranded RNA plasmid of yeast. Cell 21, 217–226 [DOI] [PubMed] [Google Scholar]
- 21. Wickner R. B. (1983) Killer systems in Saccharomyces cerevisiae: three distinct modes of exclusion of M2 double-stranded RNA by three species of double-stranded RNA, M1, L-A-E, and L-A-HN. Mol. Cell. Biol. 3, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Esteban R., Wickner R. B. (1986) Three different M1 RNA-containing virus-like particle types in Saccharomyces cerevisiae: in vitro M1 double-stranded RNA synthesis. Mol. Cell. Biol. 6, 1552–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tercero J. C., Wickner R. B. (1992) MAK3 encodes an N-acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J. Biol. Chem. 267, 20277–20281 [PubMed] [Google Scholar]
- 24. Ohtake Y., Wickner R. B. (1995) Yeast virus propagation depends critically on free 60 S ribosomal subunit concentration. Mol. Cell. Biol. 15, 2772–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edskes H. K., Ohtake Y., Wickner R. B. (1998) Mak21p of Saccharomyces cerevisiae, a homolog of human CAATT-binding protein, is essential for 60 S ribosomal subunit biogenesis. J. Biol. Chem. 273, 28912–28920 [DOI] [PubMed] [Google Scholar]
- 26. Masison D. C., Blanc A., Ribas J. C., Carroll K., Sonenberg N., Wickner R. B. (1995) Decoying the cap− mRNA degradation system by a double-stranded RNA virus and poly(A)− mRNA surveillance by a yeast antiviral system. Mol. Cell. Biol. 15, 2763–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Widner W. R., Wickner R. B. (1993) Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol. Cell. Biol. 13, 4331–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Toh-e A., Guerry P., Wickner R. B. (1978) Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 136, 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ridley S. P., Sommer S. S., Wickner R. B. (1984) Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol. Cell. Biol. 4, 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sommer S. S., Wickner R. B. (1987) Gene disruption indicates that the only essential function of the SKI8 chromosomal gene is to protect Saccharomyces cerevisiae from viral cytopathology. Virology 157, 252–256 [DOI] [PubMed] [Google Scholar]
- 31. Benard L., Carroll K., Valle R. C., Wickner R. B. (1998) Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60 S ribosomal subunit biogenesis. Mol. Cell. Biol. 18, 2688–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Searfoss A. M., Wickner R. B. (2000) 3′-Poly(A) is dispensable for translation. Proc. Natl. Acad. Sci. U.S.A. 97, 9133–9137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fujimura T., Wickner R. B. (1989) Reconstitution of template-dependent in vitro transcriptase activity of a yeast double-stranded RNA virus. J. Biol. Chem. 264, 10872–10877 [PubMed] [Google Scholar]
- 34. Tortorici M. A., Broering T. J., Nibert M. L., Patton J. T. (2003) Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J. Biol. Chem. 278, 32673–32682 [DOI] [PubMed] [Google Scholar]
- 35. Icho T., Wickner R. B. (1989) The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J. Biol. Chem. 264, 6716–6723 [PubMed] [Google Scholar]
- 36. Fujimura T., Wickner R. B. (1988) Gene overlap results in a viral protein having an RNA-binding domain and a major coat protein domain. Cell 55, 663–671 [DOI] [PubMed] [Google Scholar]
- 37. Dinman J. D., Icho T., Wickner R. B. (1991) A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. U.S.A. 88, 174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dinman J. D., Wickner R. B. (1994) Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered −1 ribosomal frameshifting efficiencies. Genetics 136, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dinman J. D., Wickner R. B. (1995) 5 S rRNA is involved in fidelity of translational reading frame. Genetics 141, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dinman J. D., Berry M. J. (2006) in Translational Control in Biology and Medicine (Mathews M. B., Sonenberg N., Hershey J. W. B., eds) pp. 625–654, Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- 41. Belew A. T., Advani V. M., Dinman J. D. (2011) Endogenous ribosomal frameshift signals operate as mRNA-destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 39, 2799–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Castón J. R., Trus B. L., Booy F. P., Wickner R. B., Wall J. S., Steven A. C. (1997) Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J. Cell Biol. 138, 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Naitow H., Tang J., Canady M., Wickner R. B., Johnson J. E. (2002) L-A virus at 3.4 Å resolution reveals particle architecture and mRNA-decapping mechanism. Nat. Struct. Biol. 9, 725–728 [DOI] [PubMed] [Google Scholar]
- 44. Leibowitz M. J., Wickner R. B. (1976) A chromosomal gene required for killer plasmid expression, mating, and spore maturation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 73, 2061–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Julius D., Brake A., Blair L., Kunisawa R., Thorner J. (1984) Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-α-factor. Cell 37, 1075–1089 [DOI] [PubMed] [Google Scholar]
- 46. Steiner D. F., Smeekens S. P., Ohagi S., Chan S. J. (1992) The new enzymology of precursor-processing endoproteases. J. Biol. Chem. 267, 23435–23438 [PubMed] [Google Scholar]
- 47. Cox B. S. (1965) Ψ, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20, 505–521 [Google Scholar]
- 48. Ono B. I., Stewart J. W., Sherman F. (1979) Yeast UAA suppressors effective in ψ+ strains serine-inserting suppressors. J. Mol. Biol. 128, 81–100 [DOI] [PubMed] [Google Scholar]
- 49. Lacroute F. (1971) Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 106, 519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aigle M., Lacroute F. (1975) Genetic aspects of [URE3], a non-mitochondrial, cytoplasmically inherited mutation in yeast. Mol. Gen. Genet. 136, 327–335 [DOI] [PubMed] [Google Scholar]
- 51. Alper T., Cramp W. A., Haig D. A., Clarke M. C. (1967) Does the agent of scrapie replicate without nucleic acid? Nature 214, 764–766 [DOI] [PubMed] [Google Scholar]
- 52. Griffith J. S. (1967) Self-replication and scrapie. Nature 215, 1043–1044 [DOI] [PubMed] [Google Scholar]
- 53. Prusiner S. B. (1982) Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 54. Singh A., Helms C., Sherman F. (1979) Mutation of the non-Mendelian suppressor, Ψ+, in yeast by hypertonic media. Proc. Natl. Acad. Sci. U.S.A. 76, 1952–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lund P. M., Cox B. S. (1981) Reversion analysis of [psi−] mutations in Saccharomyces cerevisiae. Genet. Res. 37, 173–182 [DOI] [PubMed] [Google Scholar]
- 56. Tuite M. F., Mundy C. R., Cox B. S. (1981) Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics 98, 691–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Westaway D., DeArmond S. J., Cayetano-Canlas J., Groth D., Foster D., Yang S. L., Torchia M., Carlson G. A., Prusiner S. B. (1994) Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell 76, 117–129 [DOI] [PubMed] [Google Scholar]
- 58. Chernoff Y. O., Derkach I. L., Inge-Vechtomov S. G. (1993) Multicopy SUP35 gene induces de novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 24, 268–270 [DOI] [PubMed] [Google Scholar]
- 59. Doel S. M., McCready S. J., Nierras C. R., Cox B. S. (1994) The dominant PNM2− mutation which eliminates the ψ factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 137, 659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Inge-Vechtomov S. G., Andrianova V. M. (1970) Recessive super-suppressor in yeast. Genetika 6, 103–115 [Google Scholar]
- 61. Hawthorne D. C., Leupold U. (1974) Suppressors in yeast. Curr. Top. Microbiol. Immunol. 64, 1–47 [DOI] [PubMed] [Google Scholar]
- 62. Coustou V., Deleu C., Saupe S., Begueret J. (1997) The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. U.S.A. 94, 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W. (2001) Prions affect the appearance of other prions: the story of [PIN+]. Cell 106, 171–182 [DOI] [PubMed] [Google Scholar]
- 64. Roberts B. T., Wickner R. B. (2003) Heritable activity: a prion that propagates by covalent autoactivation. Genes Dev. 17, 2083–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Du Z., Park K. W., Yu H., Fan Q., Li L. (2008) Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 40, 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Patel B. K., Gavin-Smyth J., Liebman S. W. (2009) The yeast global transcriptional corepressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 11, 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Masison D. C., Wickner R. B. (1995) Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270, 93–95 [DOI] [PubMed] [Google Scholar]
- 68. Masison D. C., Maddelein M. L., Wickner R. B. (1997) The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc. Natl. Acad. Sci. U.S.A. 94, 12503–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ter-Avanesyan M. D., Dagkesamanskaya A. R., Kushnirov V. V., Smirnov V. N. (1994) The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137, 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kirschner D. A., Teplow D. B., Damas A. M. (2000) Twist and sheet: variations on the theme of amyloid. J. Struct. Biol. 130, 87. [DOI] [PubMed] [Google Scholar]
- 71. Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268, 880–884 [DOI] [PubMed] [Google Scholar]
- 72. Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15, 3127–3134 [PMC free article] [PubMed] [Google Scholar]
- 73. King C. Y., Tittmann P., Gross H., Gebert R., Aebi M., Wüthrich K. (1997) Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. U.S.A. 94, 6618–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Glover J. R., Kowal A. S., Schirmer E. C., Patino M. M., Liu J. J., Lindquist S. (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89, 811–819 [DOI] [PubMed] [Google Scholar]
- 75. Edskes H. K., Gray V. T., Wickner R. B. (1999) The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. U.S.A. 96, 1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Taylor K. L., Cheng N., Williams R. W., Steven A. C., Wickner R. B. (1999) Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283, 1339–1343 [DOI] [PubMed] [Google Scholar]
- 77. Maddelein M. L., Dos Reis S., Duvezin-Caubet S., Coulary-Salin B., Saupe S. J. (2002) Amyloid aggregates of the HET-s prion protein are infectious. Proc. Natl. Acad. Sci. U.S.A. 99, 7402–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. King C. Y., Diaz-Avalos R. (2004) Protein-only transmission of three yeast prion strains. Nature 428, 319–323 [DOI] [PubMed] [Google Scholar]
- 79. Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S. (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 [DOI] [PubMed] [Google Scholar]
- 80. Brachmann A., Baxa U., Wickner R. B. (2005) Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24, 3082–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Patel B. K., Liebman S. W. (2007) “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p(132–405) induces [PIN+]. J. Mol. Biol. 365, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jones E. W. (1991) Three proteolytic systems in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 266, 7963–7966 [PubMed] [Google Scholar]
- 83. Zubenko G. S., Park F. J., Jones E. W. (1982) Genetic properties of mutations at the PEP4 locus in Saccharomyces cerevisiae. Genetics 102, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Priola S. A., Caughey B., Race R. E., Chesebro B. (1994) Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J. Virol. 68, 4873–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Prusiner S. B., Scott M., Foster D., Pan K. M., Groth D., Mirenda C., Torchia M., Yang S. L., Serban D., Carlson G. A., Hoppe P. C., Westaway D., DeArmond S. J. (1990) Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63, 673–686 [DOI] [PubMed] [Google Scholar]
- 86. Kochneva-Pervukhova N. V., Paushkin S. V., Kushnirov V. V., Cox B. S., Tuite M. F., Ter-Avanesyan M. D. (1998) Mechanism of inhibition of Ψ+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J. 17, 5805–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ross E. D., Baxa U., Wickner R. B. (2004) Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 24, 7206–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ross E. D., Edskes H. K., Terry M. J., Wickner R. B. (2005) Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. U.S.A. 102, 12825–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Toombs J. A., McCarty B. R., Ross E. D. (2010) Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 30, 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Toombs J. A., Petri M., Paul K. R., Kan G. Y., Ross E. D. (2012) De novo design of synthetic prion domains. Proc. Natl. Acad. Sci. U.S.A., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ross E. D., Minton A., Wickner R. B. (2005) Prion domains: sequences, structures, and interactions. Nat. Cell Biol. 7, 1039–1044 [DOI] [PubMed] [Google Scholar]
- 92. Antzutkin O. N., Balbach J. J., Leapman R. D., Rizzo N. W., Reed J., Tycko R. (2000) Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of β-sheets in Alzheimer β-amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 97, 13045–13050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Benzinger T. L., Gregory D. M., Burkoth T. S., Miller-Auer H., Lynn D. G., Botto R. E., Meredith S. C. (1998) Propagating structure of Alzheimer β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc. Natl. Acad. Sci. U.S.A. 95, 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Shewmaker F., Wickner R. B., Tycko R. (2006) Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. U.S.A. 103, 19754–19759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Baxa U., Wickner R. B., Steven A. C., Anderson D. E., Marekov L. N., Yau W. M., Tycko R. (2007) Characterization of β-sheet structure in Ure2p1–89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry 46, 13149–13162 [DOI] [PubMed] [Google Scholar]
- 96. Wickner R. B., Dyda F., Tycko R. (2008) Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc. Natl. Acad. Sci. U.S.A. 105, 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shewmaker F., Kryndushkin D., Chen B., Tycko R., Wickner R. B. (2009) Two prion variants of Sup35p have in-register parallel β-sheet structures, independent of hydration. Biochemistry 48, 5074–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shewmaker F., Ross E. D., Tycko R., Wickner R. B. (2008) Amyloids of shuffled prion domains that form prions have a parallel in-register β-sheet structure. Biochemistry 47, 4000–4007 [DOI] [PubMed] [Google Scholar]
- 99. Baxa U., Taylor K. L., Wall J. S., Simon M. N., Cheng N., Wickner R. B., Steven A. (2003) Architecture of Ure2p prion filaments. The N-terminal domains form a central core fiber. J. Biol. Chem. 278, 43717–43727 [DOI] [PubMed] [Google Scholar]
- 100. Diaz-Avalos R., King C. Y., Wall J., Simon M., Caspar D. L. (2005) Strain-specific morphologies of yeast prion amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 102, 10165–10170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chen B., Thurber K. R., Shewmaker F., Wickner R. B., Tycko R. (2009) Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 106, 14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wasmer C., Lange A., Van Melckebeke H., Siemer A. B., Riek R., Meier B. H. (2008) Amyloid fibrils of the HET-s(218–289) prion form a β-solenoid with a triangular hydrophobic core. Science 319, 1523–1526 [DOI] [PubMed] [Google Scholar]
- 103. Wickner R. B., Shewmaker F., Kryndushkin D., Edskes H. K. (2008) Protein inheritance (prions) based on parallel in-register β-sheet amyloid structures. BioEssays 30, 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wickner R. B., Shewmaker F., Edskes H., Kryndushkin D., Nemecek J., McGlinchey R., Bateman D., Winchester C. L. (2010) Prion amyloid structure explains templating: how proteins can be genes. FEMS Yeast Res. 10, 980–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wickner R. B., Edskes H. K., Kryndushkin D., McGlinchey R., Bateman D., Kelly A. (2011) Prion diseases of yeast: amyloid structure and biology. Semin. Cell Dev. Biol. 22, 469–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wickner R. B., Edskes H. K., Shewmaker F., Nakayashiki T. (2007) Prions of fungi: inherited structures and biological roles. Nat. Rev. Microbiol. 5, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wickner R. B. (1997) A new prion controls fungal cell fusion incompatibility. Proc. Natl. Acad. Sci. U.S.A. 94, 10012–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Dalstra H. J., Swart K., Debets A. J., Saupe S. J., Hoekstra R. F. (2003) Sexual transmission of the [Het-S] prion leads to meiotic drive in Podospora anserina. Proc. Natl. Acad. Sci. U.S.A. 100, 6616–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nakayashiki T., Kurtzman C. P., Edskes H. K., Wickner R. B. (2005) Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. U.S.A. 102, 10575–10580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jung G., Jones G., Wegrzyn R. D., Masison D. C. (2000) A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156, 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Schwimmer C., Masison D. C. (2002) Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22, 3590–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hosoda N., Kobayashi T., Uchida N., Funakoshi Y., Kikuchi Y., Hoshino S., Katada T. (2003) Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278, 38287–38291 [DOI] [PubMed] [Google Scholar]
- 113. Hoshino S., Imai M., Kobayashi T., Uchida N., Katada T. (1999) The eukaryotic polypeptide chain-releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 274, 16677–16680 [DOI] [PubMed] [Google Scholar]
- 114. Funakoshi Y., Doi Y., Hosoda N., Uchida N., Osawa M., Shimada I., Tsujimoto M., Suzuki T., Katada T., Hoshino S. (2007) Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 21, 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shewmaker F., Mull L., Nakayashiki T., Masison D. C., Wickner R. B. (2007) Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 176, 1557–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Edskes H. K., Engel A., McCann L. M., Brachmann A., Tsai H. F., Wickner R. B. (2011) Prion-forming ability of Ure2 of yeasts is not evolutionarily conserved. Genetics 188, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Engel A., Shewmaker F., Edskes H. K., Dyda F., Wickner R. B. (2011) Amyloid of the Candida albicans Ure2p prion domain is infectious and has an in-register parallel β-sheet structure. Biochemistry 50, 5971–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Safadi R. A., Talarek N., Jacques N., Aigle M. (2011) Yeast prions: could they be exaptations? The URE2/[URE3] system in Kluyveromyces lactis. FEMS Yeast Res. 11, 151–153 [DOI] [PubMed] [Google Scholar]
- 119. Edskes H. K., McCann L. M., Hebert A. M., Wickner R. B. (2009) Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics 181, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Parry H. B. (1983) Scrapie disease in Sheep: Historical, Clinical, Epidemiological, Pathological and Practical Aspects of the Natural Disease, Academic Press, London [Google Scholar]
- 121. Wickner R. B. (2005) Scrapie in Ancient China? Science 309, 874. [DOI] [PubMed] [Google Scholar]
- 122. Hunter N. (1997) PrP genetics in sheep and the applications for scrapie and BSE. Trends Microbiol. 5, 331–334 [DOI] [PubMed] [Google Scholar]
- 123. Mead S., Stumpf M. P., Whitfield J., Beck J. A., Poulter M., Campbell T., Uphill J. B., Goldstein D., Alpers M., Fisher E. M., Collinge J. (2003) Balancing selection at the prion protein gene consistent with prehistoric kuru-like epidemics. Science 300, 640–643 [DOI] [PubMed] [Google Scholar]
- 124. Bateman D. A., Wickner R. B. (2012) [PSI+] prion transmission barriers protect Saccharomyces cerevisiae from infection: intraspecies ‘species barriers.’ Genetics 190, 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. McGlinchey R. P., Kryndushkin D., Wickner R. B. (2011) Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. U.S.A. 108, 5337–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kirkland P. A., Reidy M., Masison D. C. (2011) Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 188, 565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Westermark G. T., Westermark P. (2010) Prion-like aggregates: infectious agents in human disease. Trends. Mol. Med. 16, 501–507 [DOI] [PubMed] [Google Scholar]
- 128. Moreno-Gonzalez I., Soto C. (2011) Misfolded protein aggregates: mechanisms, structures, and potential for disease transmission. Semin. Cell Dev. Biol. 22, 482–487 [DOI] [PMC free article] [PubMed] [Google Scholar]