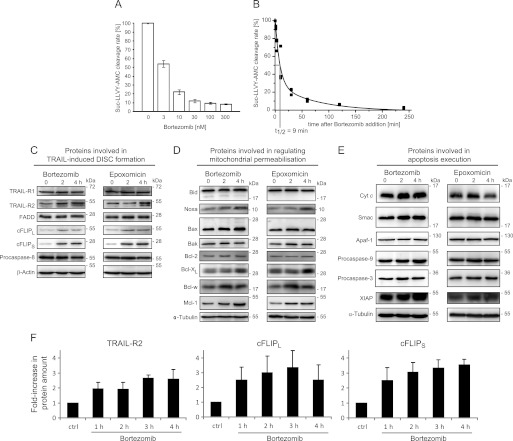
FIGURE 1.
Biochemical analysis of the impact of proteasome inhibition on the protein composition of the apoptotic TRAIL-signaling pathway in HeLa cells. A, HeLa cells were treated with the indicated concentrations of bortezomib for 4 h, and chymotrypsin-like proteasome activity was measured by cleavage of Suc-LLVY-AMC. Maximal inhibition was achieved with doses ≥100 nm. Remaining residual Suc-LLVY-AMC cleavage was likely due to other enzymes or substrate hydrolysis. Data are shown as mean ± S.E. from eight samples pooled from two independent experiments. B, HeLa cells were treated with 100 nm bortezomib, and Suc-LLVY-AMC cleavage rates were fitted to a biphasic exponential decay function following background subtraction. Data were pooled from n = 3 independent experiments with triplicates. C–E, whole cell extracts of HeLa cells treated with 100 nm bortezomib or 50 nm epoxomicin for the indicated times were analyzed by immunoblotting. β-Actin or α-tubulin served as loading controls. Experiments were repeated with similar results. C, proteins involved in DISC formation and caspase-8 activation. D, proteins regulating mitochondrial outer membrane permeabilization. E, proteins involved in apoptosis execution. F, quantitative analysis of TRAIL-R2 and cFLIP species accumulation in response to 100 nm bortezomib. Protein amounts were quantified by immunoblot densitometry. Data are means + S.E. from n = 4 independent experiments.