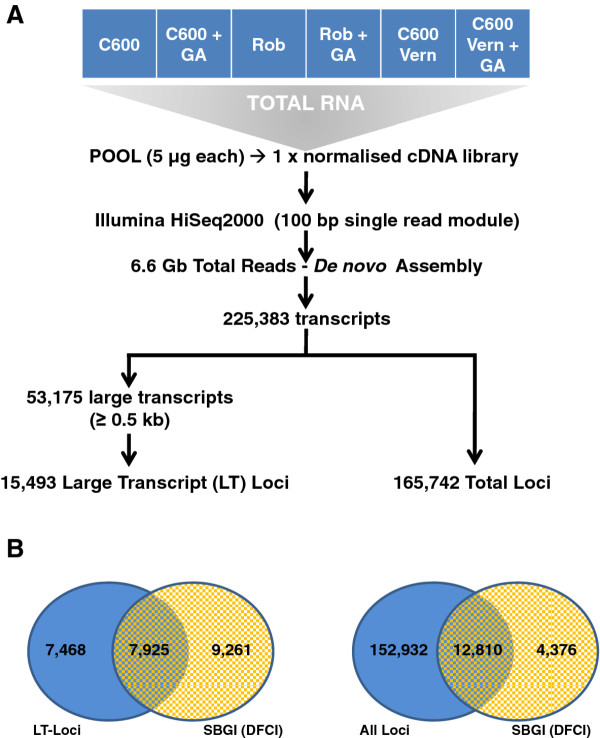
Figure 2.
Overview of the reference transcriptome sequencing, and result of the Velvet/Oases assembly. A) RNA for the reference was pooled from all of the test samples. The Illumina HiSeq2000 platform was used to generate data for de novo assembly including large transcripts (LT) equivalent to 15,493 potential protein coding sequences. B) The accuracy and integrity of the assembly was assessed by BLAST comparison (100 bp overlap and ≥ 98% sequence identity) with the publicly available collection of sugar beet ESTs at the Sugar Beet Gene Index (SBGI), hosted at the Dana Farber Cancer Institute (DFCI).