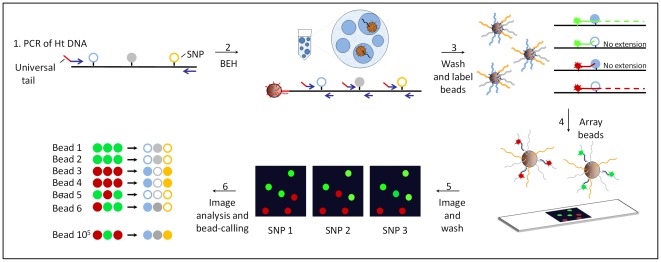
Figure 1. Schematics of bead-emulsion haplotyping (BEH).
In step 1, a region of several kilobases containing several polymorphic sites is amplified from genomic DNA heterozygous for the tested SNPs. In step 2, single amplicons are hybridized to microscopic beads covered with a primer that has a universal tail common to the 5′ end of the multiplexing primers (shown in red). PCR products from 3 small regions containing the polymorphisms are produced within the aqueous compartment of an emulsion droplet and bound to the bead. In step 3, the beads are washed and labeled by allele-specific extensions of fluorescent probes specific for one of the polymorphic sites. In step 4, unextended probes are washed off and the fluorescent beads are arrayed on a slide. In step 5, the array is scanned with a microscope followed by subsequent washing, probing and imaging cycles to screen additional polymorphisms (up to 4 different alleles; 2SNPs can be analyzed simultaneously). In step 6, a series of imaging and data analysis steps are performed to assess the initial haplotype of ∼105 molecules.