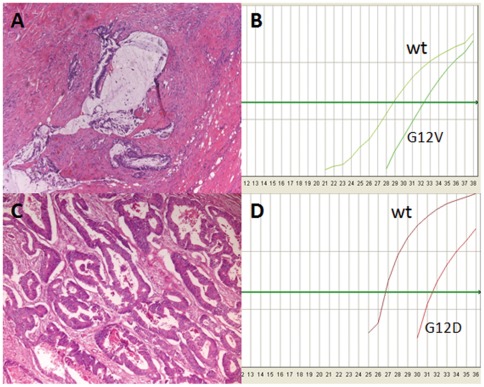
Figure 6. KRAS mutations identified by ASLNAqPCR but not by Sanger sequencing. A.
, Hematoxylin and Eosin (H&E) stained section (X100) of the area of case 13 of Table 7 (rectal adenocarcinoma treated with preoperative chemo– and radiation therapy) dissected for DNA extraction with a tumor vs. non neoplastic cell ratio of ∼10%, below the analytical sensitivity threshold of Sanger sequencing. B, ASLNAqPCR of case 13 of Table 7 shows a KRAS G12V mutation with a mutated/wild type ratio of 4%, corresponding to 8% mutated cells, assuming that the mutation is heterozygous; this is consistent with the mutation being present in the large majority of neoplastic cells. C, H&E stained section (X100) of the area of the colonic adenocarcinoma case 1 of Table 7, dissected for DNA extraction with a tumor vs. non neoplastic cell ratio of ∼70%. D, ASLNAqPCR of case 1 of Table 7 shows a KRAS G12D mutation with a mutated/wild type ratio of 1.5%, corresponding to 3% mutated cells, assuming that the mutation is heterozygous; this is consistent with a small KRAS G12D mutated subclone corresponding to ∼4% of the neoplastic cells.